Genes & Cancer
Signaling transcript profile of the asexual intraerythrocytic development cycle of Plasmodium falciparum induced by melatonin and cAMP
Wânia Rezende Lima1,6,*, Giulliana Tessarin-Almeida1,*, Andrei Rozanski2, Kleber S. Parreira3, Miriam S. Moraes1, David C. Martins4, Ronaldo F. Hashimoto5, Pedro A.F. Galante2 and Célia R.S. Garcia1
1 Departamento de Fisiologia, Instituto de Biociências, Universidade de Sao Paulo, Sao Paulo, Brazil
2 Centro de Oncologia Molecular, Hospital Sírio-Libanês, Sao Paulo, Brazil
3 Departamento de Imunologia e Parasitologia, Instituto de Ciências Biomédicas, Universidade Federal de Uberlândia, Brazil
4 Centro de Matemática, Computação e Cognição, Universidade Federal do ABC, São Paulo, Brazil
5 Departamento de Ciência da Computação, Instituto de Matemática e Estatística, Universidade de São Paulo, São Paulo, Brazil
6 Instituto de Ciências Exatas e Naturais (ICEN)- Medicina, Universidade Federal do Mato Grosso - Campus Rondonópolis, Brazil
* These first authors equally contributed to this work
Correspondence to: Célia R.S. Garcia, email: [email protected]
Keywords: transcripts, malaria, P. falciparum, melatonin, cAMP, ubiquitin proteasome
Received: May 31, 2016
Accepted: September 25, 2016
Published: October 03, 2016
ABSTRACT
According to the World Health Organization (WHO), Plasmodium falciparum is the deadliest parasite among all species. This parasite possesses the ability to sense molecules, including melatonin (MEL) and cAMP, and modulate its cell cycle accordingly. MEL synchronizes the development of this malaria parasite by activating several cascades, including the generation of the second messenger cAMP. Therefore, we performed RNA sequencing (RNA-Seq) analysis in P. falciparum erythrocytic stages (ring, trophozoite and schizont) treated with MEL and cAMP. To investigate the expression profile of P. falciparum genes regulated by MEL and cAMP, we performed RNA-Seq analysis in three P. falciparum strains (control, 3D7; protein kinase 7 knockout, PfPK7-; and PfPK7 complement, PfPK7C). In the 3D7 strain, 38 genes were differentially expressed upon MEL treatment; however, none of the genes in the trophozoite (T) stage PfPK7- knockout parasites were differentially expressed upon MEL treatment for 5 hours compared to untreated controls, suggesting that PfPK7 may be involved in the signaling leading to differential gene expression. Moreover, we found that MEL modified the mRNA expression of genes encoding membrane proteins, zinc ion-binding proteins and nucleic acid-binding proteins, which might influence numerous functions in the parasite. The RNA-Seq data following treatment with cAMP show that this molecule modulates different genes throughout the intraerythrocytic cycle, namely, 75, 101 and 141 genes, respectively, in the ring (R), T and schizont (S) stages. Our results highlight P. falciparum’s perception of the external milieu through the signaling molecules MEL and cAMP, which are able to drive to changes in gene expression in the parasite.
INTRODUCTION
Malaria is a major cause of death worldwide, and Plasmodium falciparum is the species responsible for the majority of malaria cases. In mammals, the parasite P. falciparum has adapted to living in red blood cells during its asexual life cycle. Our previous work showed that the host circadian rhythm, which is regulated by melatonin (MEL), a molecule produced by the pineal gland in response to darkness [1], plays an important role in controlling proliferation and synchronizing the development of the malaria parasite [2, 3]. This regulation is achieved by activating signaling cascades, including the PLC-IP3 pathway; cross-talking; and generating the second messengers cAMP and IP3, which elicit an increase in cytosolic calcium. As to the molecular machinery involved, these components could include proteases, kinases and genes that belong to the UPS [4, 5]. More strikingly, the protein kinase 7 (PK7) knockout strain PfPK7 is unable to respond to MEL and to synchronize the intraerythrocytic forms of the parasites [6].
In recent decades, much effort has been focused on understanding how gene regulation is achieved in Plasmodium. Unfortunately, although the P. falciparum genome sequencing is complete [7], half of the genes remain without annotation; therefore, understanding parasite biology poses a large challenge. Microarray technology was used to obtain information regarding the transcriptome of the parasite during its sexual development [8] and during the intraerythrocytic developmental cycle [9] to profile the malaria parasite in its different life stages [10]. Moreover, high-throughput sequencing of cDNA (RNA-Seq) presented a new opportunity to deeply probe the Plasmodium transcriptome. RNA-Seq analyses allowed the detection of novel gene transcripts, the correction of a large number of genes, and the refinement of the original gene model, revealing tight regulation of gene expression throughout the intraerythrocytic development cycle of P. falciparum [11-16].
The mechanisms for controlling the cell cycle and development in malaria parasites are far from being fully understood. Dissecting the cellular signaling networks is an important goal for not only understanding the biology of these parasites but also providing new ways to combat malaria infection [17].
In the last decade, studies from several labs considerably advanced our understanding of how the organisms in the genus Plasmodium sense the signaling milieu and trigger their cellular and molecular machinery involved in transducing these signals [4, 6, 18-21].
Recently, cAMP has been identified as a key regulator that triggers the timely secretion of microneme proteins, enabling receptor engagement and invasion [22]. This study also observed that cAMP increases merozoite cytosolic Ca2+ levels via induction of the Epac pathway; together, they are involved in the invasion process [22]. Moreover, our group reported that the cAMP analog adenosine 3’,5’-cyclic monophosphate N6-benzoyl/PKA activator (6-Bz-cAMP) is able to increase the schizont (S) stage, leading to progression of the asexual stage of the life cycle and modulation of PFNF-YB transcription factor expression during the intraerythrocytic cycle in P. falciparum [23].
However, how these parasites respond to regulation by the host molecules MEL and cAMP, which modulate the proliferation cycle in vitro, is still unknown. It is becoming increasingly clear that chromatin remodeling and epigenetic memory play an important role in the expression and antigenic variation of virulence genes in infected red blood cells (iRBCs) [24-26].
We generated RNA-Seq data to study P. falciparum-infected RBCs at the trophozoite (T) stage of the wild-type (WT) strain (3D7), the PK7 knockout strain (PfPK7-) and its complement strain (PfPK7C) to determine the target genes involved in parasite synchronization by MEL. This approach also helped us to identify possible targets and the role of PK7 in gene transcription. The results revealed that incubation of P. falciparum-infected cells with MEL for 5 hours led to considerable changes in gene expression in the 3D7 strain compared to that of the control. When similar experiments were performed in PfPK7 knockout strains, we did not observe any differential gene expression in MEL treated and untreated assays. These findings are strong evidence of the relevance of this gene (PfPK7) in MEL-induced regulation of the cell cycle. To better understand the signaling cascade activated by cAMP, we treated ring (R), T and S stage parasites with the cAMP analog 6-Bz-cAMP. Our results showed that cAMP and MEL treatment affected the mRNA transcript levels of UPS genes at the T stage. In addition to the modulation of kinases and transcription factors, cAMP treatment altered the expression of ribosomal proteins at the R stage and of isomerase, rotamase and binding proteins involved in metabolic processes, such as coenzymes, at the S stage.
RESULTS
Sequencing and data processing
We previously reported that a subset of genes related to the ubiquitin proteasome system (UPS) was modulated in P. falciparum after 5 hours of treatment with MEL [6]. Therefore, we performed RNA-Seq analysis with synchronized parasites in three P. falciparum strains (control 3D7, protein kinase 7 knockout, PfPK7-, and PfPK7 complementPfPK7C).
We collected total RNA from synchronized T stage 3D7 parasites that were treated with MEL (final concentration, 100 nM) and incubated for 5 hours at 37°C. Total RNA samples were processed as described below (see Methods and Supplementary Figure 1). Every treatment and RNA-Seq were performed on two biological replicates. In summary, ~81.6% (3D7), ~79% (PfPK7-) and ~81.5% (PfPK7C) of reads were mapped uniquely against the Plasmodium genome to verify the sequencing quality (Supplementary Tables 1 and 2).
Melatonin treatment leads to a change in P. falciparum gene expression
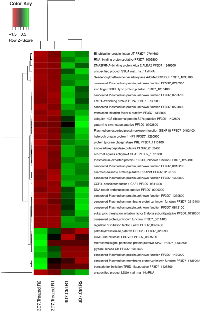
Next, we compared the gene expression profiles of the P. falciparum parasite lines (3D7 and PfPK7-clones) untreated and treated with MEL at 37°C. The results revealed that 100 nM MEL caused a change in gene expression only for the 3D7 strain after 5 hours of incubation. Among the 38 differentially expressed genes in the MEL-treated 3D7 strain, 7 genes were down-regulated and 31 genes were up-regulated (p-value < 0.05; FDR < 0.05) compared with the treated and untreated 3D7 strains (Supplementary Table 3 and Figure 1).
The differentially expressed genes up-regulated by the MEL treatment included E3 ubiquitin ligase (PF3D7_0704600); regulator of initiation factor 2 (eIF2; PF3D7_0322400), which plays an important role in post-transcriptional control of gene expression in malaria parasites; and the gene encoding S-adenosylmethionine decarboxylase/ornithine decarboxylase (AdoMetDC/ODC; PF3D7_1033100).
The MEL treatment also up-regulated a series of genes related to RNA metabolism, including a zinc finger protein (PF3D7_0202900), zinc finger (CCCH type) protein (PF3D7_1315400), translation initiation factor 6 (PF3D7_1332800), eukaryotic translation initiation factor 2 alpha subunit (PF3D7_0728000), transcription initiation TFIID-like (PF3D7_1428800), DNA/RNA-binding protein Alba 2 (ALBA2; PF3D7_1346300), RNA-binding protein (PF3D7_1006800), DNA repair endonuclease (PF3D7_0206000), ubiquitin-40S ribosomal protein S27a (PF3D7_1402500), and heterochromatin protein 1 (PF3D7_1220900).
Additionally, the gene encoding CCR4-Associated Factor 1 (CAF1) was up-regulated after the MEL treatment. CAF1 coordinates the expression of P. falciparum egression and invasion proteins, which are extremely important for asexual blood-stage development [27].Among the seven differentially expressed genes down-regulated by the MEL treatment, we found microsomal signal peptidase (PF3D7_1433600), which has an important role in the transport and maturation of parasite proteins; glycerol kinase (GK; PF3D7_1351600), which is required for optimal intraerythrocytic asexual parasite development; SPC1 (PF3D7_1433600) and RUVBL31 (PF3D7_0809700).
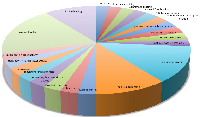
Next, we identified the gene function of these up- and down-regulated genes. Figure 2 is a pie chart showing the distribution of Gene Ontology (GO) [28] terms for the 38 differentially expressed genes in the 3D7 strain. Using GO annotation, we found that 4 of 11 genes that transcribe conserved Plasmodium proteins with unknown function are annotated as genes coding for membrane proteins, one gene is annotated as a gene coding for a nuclear protein, one gene is annotated as a gene coding for protein serine/threonine kinase activity, one gene is annotated as a gene coding for actin binding, and one gene is annotated as a gene coding for zinc ion binding. In total, 29% of the differentially expressed genes (11 genes) had no functional annotation.
cAMP stimulates cell division and represses cell host invasion gene expression in the trophozoite stage of P. falciparum
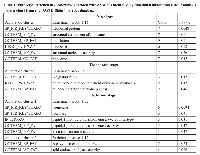
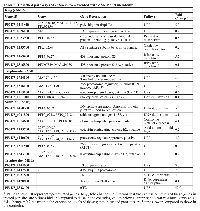
The second messenger cAMP participates in several biological activities in eukaryotic cells and interplays with the MEL pathway [5, 29]. Because 6-Bz-cAMP is able to modulate the P. falciparum life cycle by increasing the number of parasites at the S stage, thus leading to progression of the asexual stage of the life cycle, we investigated the role that cAMP plays during the asexual intraerythrocytic cycle of P. falciparum. Highly synchronous trophozoites at the 34-hours timepoint were treated with 6-Bz-cAMP compound (Supplementary Figure 2). Every treatment and RNA-Seq were performed on two biological replicates. At the T stage, we found that 49 genes were up-regulated, and 52 genes were down-regulated (Figure 3 and Supplementary Table 4). We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool to search for genes involved in the cAMP pathway in P. falciparum [30, 31]. Eighty-one DAVID gene IDs were identified, but only six chart records were found. The analysis revealed that three of six pathways noted from the functional annotation were significantly modified by cAMP (see Table 1).
cAMP signaling targets different genes during the intraerythrocytic cell cycle of P. falciparum
We also investigated the cAMP signaling targets at the R and S stages. To achieve this goal, we performed RNA-Seq using samples from highly synchronized iRBCs. The parasite strain 3D7 was synchronized in R and S forms. After the iRBCs were synchronized, they were selected at 6 hours and 44 hours for treatment with 20 µM of a cAMP analog for 6 hours at 37°C. The genome-wide transcriptome analysis of the R and S stages revealed 75 and 141 genes, respectively, that were differentially expressed based on fold changes greater than 2.0 or lower than -2.0, CPM ≥ 1, and a p-value < 0.05.
To understand the role of cAMP during the R stage, analyses of the whole transcripts modified following cAMP analog treatment were performed. We identified 36 up-regulated genes and 39 down-regulated genes (Figure 4). Using the DAVID bioinformatics database, 66 DAVID gene IDs were functionally annotated. Three clusters of biological activities were formed, and one group of ribosomal genes presented a significant p-value (Table 1). The genes in this cluster were found to be related to ribosomal structure and function as identified by GO term annotation and KEGG pathway analysis. These genes were classified in (a) ribosomal protein, (b) structural constituent of ribosome, (c) translation, (d) ribosome and (e) structural molecule activity. KEGG pathway analysis identified a variety of up-regulated genes involved in structural constituents of ribosomes, such as small subunit ribosomal proteins RP-S11e, RP-S19e, RP-S26e, and RP-S3e (PF3D7_0317600/K02949, PF3D7_0422400/K02966, PF3D7_0217800/K02976 and PF3D7_1465900/K02985); biosynthesis of secondary metabolites and antibiotics, such as pyruvate dehydrogenase E1 component beta subunit and pyrroline-5-carboxylate reductase (PF3D7_1446400/K00162 and PF3D7_1357900/K00286). By contrast, several ATP-binding proteins were found to be down-regulated upon cAMP treatment: chromodomain-helicase-DNA-binding protein 1 homolog, putative (CHD1; PF3D7_1023900); protein kinase 2 (PK2; PF3D7_1238900); carbamoyl phosphate synthetase (cpsSII, PF3D7_1308200); a conserved Plasmodium protein of unknown function (PF3D7_0315400); serine/threonine protein kinase, FIKK family (FIKK4.2; PF3D7_0424700), cdc2-related protein kinase 3 (CRK3; PF3D7_0415300); and ATP synthase subunit beta, mitochondrial (PF3D7_1235700). For additional details, see Supplementary Table 5.
The DAVID bioinformatics database was used to analyze the differentially expressed genes regulated by cAMP treatment during the S stage. In total, 132 DAVID gene IDs matched 141 genes identified as differentially expressed by RNA-Seq. Functional annotation generated ten clusters, and only two clusters presented a significant p-value (Table 1). Cluster 1 represented isomerase activity, and cluster 2 represented coenzyme metabolic process and acid-amino acid ligase activity genes. Among the genes coding for isomerases and rotamases, cAMP modulated genes coding for the following peptidyl-prolyl cis-trans isomerases: CYP81 (PF08_0128), CYP72 (PFI1490c), and CYP32 (PFL0375w) and triosephosphate isomerase (TIM; PF14_0378). In cluster 2, coenzyme metabolic process was modulated by mRNA expression of dihydrofolate synthase/folylpolyglutamate synthase (DHFS-FPGS; PF13_0140), glutathione synthetase (GS; PFE0605c), ribose 5-phosphate epimerase (PFE0730c), and gamma-glutamylcysteine synthetase (gammaGCS; PFI0925w). Furthermore, the following genes involved in acid-amino acid ligase activity were down-regulated by cAMP (Figure 5): DHFS-FPGS (PF13_0140), GS (PFE0605c), gammaGCS (PFI0925w) and ubiquitin-conjugating enzyme E2 (PFL2100w).
We also observed a series of up- and down-regulated kinases (Figure 5 and Supplementary Table 6). Many Plasmodium kinases have recently been shown by reverse genetics to be crucial for various parts of the complex parasitic life cycle and are therefore potential targets [32]. Among the up-regulated kinases, we would like to highlight serine/threonine protein kinase, FIKK family (FIKK10.2; PF3D7_1039000); protein kinase (PF3D7_0926300); and nucleoside diphosphate kinase (NDK; PF3D7_1366500).
Recently, data mining of the P. falciparum genome revealed a new putative kinase family, which was called the FIKK family because of the conservation of a phenylalanine, isoleucine, lysine, and lysine amino acid motif [33-35]. The FIKK gene is located at chromosomal subtelomeric regions near several other differentially expressed genes that are important for remodeling the intraerythrocytic membrane [36]; consequently, FIKK could be involved in specific host-parasite interactions, such as the regulation of expression of the var gene family [36]. Nucleoside diphosphate kinase plays a crucial role in maintaining the intracellular energy resources and balancing nucleotide pools. [37]. Nucleoside diphosphate kinase plays a unique metabolic role, as this enzyme performs the last reaction of ribonucleotide and deoxyribonucleotide biosynthesis. This enzyme was found to bind a broad range of ribo- and deoxyribonucleotides; thus, it can produce all precursors of DNA and RNA synthesis. NDK produces NTP for nucleic acid synthesis, CTP for lipid synthesis, UTP for polysaccharide synthesis and GTP for protein synthesis, signal transduction or microtubule polymerization [38].
Among the down-regulated kinases, we highlight protein serine/threonine kinase-1 (CLK1; PF3D7_1445400), cdc2-related protein kinase 3 (CRK3; PF3D7_0415300) and cdc2-related protein kinase 5 (CRK5; PF3D7_0615500). CLK1 is part of the CMGC group of kinases, which interact with factors involved in mRNA splicing. As previously described, PfLAMMER (PF14_0431) [39] is related to yeast kns1 [40] and the human LAMMER kinases CLK1-4, which also phosphorylate SR proteins [41]. CRK3 encodes an uncommonly large CDK-related protein (1,339 amino acids) whose kinase domain displays maximal homology to those CDKs, and this protein is involved in the control of transcription in other eukaryote organisms [35]. It has been hypothesized that PfCRK-3 is part of a complex containing other protein kinases because this enzyme is associated with a kinase activity present in the parasite extracts. Moreover, it was previously demonstrated that Pfcrk-3 interacts with a histone deacetylase (HDAC) in parasite extracts, suggesting that the pfcrk-3 gene plays a crucial role in parasite proliferation during the asexual erythrocytic cycle [35].
DISCUSSION
Decoding how transcription is modulated in P. falciparum is fundamental to understanding parasite cell cycle regulation. MEL is a ubiquitous molecule that plays a central role in plants, mammals and parasites [42]. Here, we used the WT 3D7 and PfPK7- strains of P. falciparum to investigate transcriptional changes at various developmental stages upon MEL treatment. We attempted to complement the function of PfPK7 by episomal expression of the gene; however, this parasite strain did not show a complementation phenotype (data available at the online data repository) possibly due to incorrect timing of expression or low expression levels from the episomal plasmid. The identified E3 ligase (PF3D7_0704600) is likely part of a ubiquitin-mediated pathway that is triggered by an elegant activation and transfer cascade [43]. Once MEL and the UPS interact, gene expression of UPS members in P. falciparum is temporarily controlled [44](41); it is extremely important to identify these genes among the differentially expressed genes following MEL treatment. Interestingly, MEL and UPS also interact in different biological systems [45].
Another up-regulated gene is the gene encoding S-adenosylmethionine decarboxylase/ornithine decarboxylase (AdoMetDC/ODC; PF3D7_1033100). Polyamine synthesis is mainly under the control of the rate-limiting decarboxylases ODC (ornithine decarboxylase) and AdoMetDC (S-adenosylmethionine decarboxylase) [46](43). The intermolecular interactions between AdoMetDC and ODC seem to be vital for optimal ODC activity [47-49](44-46) and seem to be a viable strategy to aid in the design of selective inhibitors of polyamine metabolism of P. falciparum [50](47). The protein ALBA (Acetylation lowers binding affinity; PF3D7_1346300) is part of the DNA/RNA-binding protein family [51, 52](48,49). MEL treatment also causes the up-regulation of a gene encoding an RNA-binding protein. Recently, the protein encoded by this gene was identified as a homolog of Gbp2 [53]. Gbp2 seems to be expressed in all the developmental stages of the parasite (based on PlasmoDB expression data). The homolog of Gbp2, PfGbp2, is a small protein containing an arginine-rich region and two RNA recognition motifs (RRMs), one in the N-terminal and the other in the C-terminal region of the protein.
The proteins known as eukaryotic initiation factors (eIFs) play an important role in post-transcriptional control of gene expression in malaria parasites [54](52); their expression results in the selective translation of mRNAs encoding stress-response proteins. We observed up-regulation of the gene coding eIF2 (PF3D7_0322400) in the T stage parasites treated with MEL.
The P. falciparum heterochromatin protein 1 (PF3D7_1220900) was identified as a major structural component of virulence gene islands throughout the parasite genome; it binds specifically to a reversible histone modification, which marks these virulence loci for transcriptional silencing [55](53). Additionally, HP1 was identified as a protein that controls the antigenic variation of a protozoan parasite [56](54). MEL treatment up-regulated two genes coding for Plasmodium exported proteins GEXP18 (PF3D7_0402400) and PHISTb (PF3D7_1252800), which could indicate a unifying epigenetic strategy in the regulation of host-parasite interactions and immune evasion in P. falciparum.
The differentially expressed genes that were down-regulated by MEL treatment also included microsomal signal peptidase protein (PF3D7_1433600) and glycerol kinase (GK) (PF3D7_1351600). Signal peptidases are membrane-bound endopeptidases that have an important role in the transport and maturation of parasite proteins [57](55).
PfSEA1 (PF3D7_1021800), essential for blood-stage replication [58],(58) and protein tyrosine phosphate PfPRL (PF3D7_1113100), which plays a role in the merozoite stage, were positively modulated by MEL.
We investigated the effect of cAMP on the R, T, and S stages of the parasite life cycle, which lasts 48 hours. For each stage, we selected one point: R (6 hours), T(34 hours) and S(44 hours). Our results showed that cAMP could modulate several genes implicated in a variety of biological activities. Specifically, at the T stage following cAMP treatment for 6 hours, genes involved in cell division, proliferation, egress and UPS were up-regulated. Moreover, metabolic pathways such as fatty acid biosynthesis and degradation, spliceosome, peroxisome, citrate cycle and ubiquinone were positively modulated. The KROX1 protein is associated with proliferation [61], and the 26S proteasome regulatory subunit RPN3 and anaphase promoting complex subunit are crucial for UPS operation. Indeed, we believe that the cross-talk between MEL signaling and cAMP partially occurs through UPS proteins because they are also down-regulated by cAMP treatment. Examples of these UPS proteins include ubiquitin protein ligase, putative (HRD3) and ubiE/COQ5 methyltransferase. MEL treatment augmented E3 ubiquitin ligase and ubiquitin-40S ribosomal protein S27a mRNA levels, showing that they did not regulate the same genes. However, cAMP and MEL share the same signaling pathway that controls protein activities and, consequently, the cell cycle and cell development [4, 5]; therefore, we presented these results in Table 2. MEL and cAMP modulate genes that are coupled to aerobic respiration and ATP production, such as S-adenosylmethionine decarboxylase/ornithine decarboxylase (AdoMetDC/ODC), glutamine synthetase, succinyl-CoA synthetase alpha subunit and acyl-CoA synthetase (ACS11). These results may explain why the parasite replicates faster when treated with MEL and cAMP; the regulation of these groups of genes favors metabolic pathways, thus accelerating parasite maturation.
The NIMA-related serine/threonine kinases NEK3 mRNA expression levels increased due to cAMP treatment. NEK3 belongs to the NEK family of proteins, which are important for DNA replication and which have been implicated as possible regulators of an atypical mitogen-activated protein kinase (MAPK) because both enzymes synergize with Pfmap-2 MAPK in vitro [62].
cAMP treatment of infected RBCs led to an increase in the proportion of schizonts; we found that an increase in the mRNA expression of genes involved in the cell cycle and DNA replication, such as RPA1 [63], SEA1, NDK and the myb2 transcription factor. The up-regulation of this set of genes partially explains how the cAMP-treated parasite maintains a large number of schizonts. RPA1 gene activity correlates with the timing of chromosomal DNA replication [63]. Antibodies against PfSEA1 decreased parasite replication and blocked schizont rupture [60], suggesting that this protein is important for merozoite egression. SAS-4 protein is exclusively expressed in the late S stage [64]. Myb proteins bind DNA in a sequence-specific manner and regulates the expression of genes involved in differentiation and growth control [65].
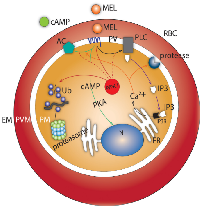
cAMP negatively modulates genes involved in the UPS and protein degradation,such as E2 ubiquitin, 26S proteasome, APC and falstatin. Likewise, in T and S stages, cAMP also regulates UPS genes. These data suggest that cAMP may modulate UPS genes independent of the activation of a MEL-mediated signaling cascade. Not only UPS genes but also genes involved in the cell cycle, cell invasion and egress appear to be modulated independent of the MEL cascade. Figure 6 shows a schematic of MEL and cAMP action during the P. falciparum asexual stage of development. Our results suggest that genes involved in replication, release and merozoite invasion, such as CCR4, which is crucial for the regulation of proteins involved in egression and invasion events, may be excellent candidates for controlling parasite proliferation and for designing new antimalarial drugs.
MATERIALS AND METHODS
Plasmodium falciparum culture
Plasmodium falciparum parasite lines (3D7, PfPK7-and PfPK7C clones) were cultured in 25 cm² flasks at 37°C and 5% hematocrit in RPMI 1640 medium supplemented with 10% human plasma, gassed with 90% N2, 5% O2 and 5% CO2. The synchronous parasite was obtained from R stage parasites, incubated for 10 minutes with 5 % sorbitol solution culture, and centrifuged for 5 minutes at 800 × g at room temperature. The supernatant was discarded, and the erythrocytes were washed twice with complete RMPI medium and added back to the normal culture as previously described[66]. For PfPK7- and PfPK7C parasites, the same medium was supplemented with 2.5 μg/mL blasticidin.
Parasite synchronization
The P. falciparum 3D7 strain culture was highly synchronized as follows: the cultured parasites that had reached the R stage were centrifuged at ~300 × g for 5 minutes at room temperature. The medium was discarded, and parasite-infected RBCs were incubated with 5% sorbitol for 10 minutes. After treatment, the culture was centrifuged again, and the sorbitol was removed. The culture was washed twice with PBS, and RPMI medium was added [67]. The knockout strain PfPK7 was kindly provided by Professor Christian Doerig [68].
Melatonin treatment
The cultured erythrocytes previously synchronized with parasites were transferred to a large flask, and fresh medium was added. MEL (final concentration, 100 nM) was immediately added to the synchronized parasites in culture at the T stage (30 hours after infection) and incubated for 5 hours [6] at 37°C. Every treatment was performed on two biological replicates.
6-BnZ-cAMP analog treatment
All experiments were performed using human erythrocytes. The culture of erythrocytes infected with P. falciparum was highly synchronized using sorbitol. Intraerythrocytic development cycle of P. falciparum was monitored by smear and Giemsa staining. Malaria parasites have a 48-hour asexual cycle inside RBCs; thus, the selected hours were representative of the 3 stages (R, T, and S). R, T, and S parasites at 6 hours, 34 hours and 44 hours, respectively, were used for cAMP assays. At each timepoint, 20 µM 6-Bnz-cAMP analog was added, and the cultures were incubated for 6 hours at 37°C. Every treatment was performed on two biological replicates.
RNA extraction and library construction
Total RNA extraction was performed following the protocol of Ponts et al. [69], with some modifications. Parasites from highly synchronous cultures were harvested at 12, 30, 40 and 50 hours post-infection and after MEL and cAMP treatments. The cultures were centrifuged at 700 × g for 5 minutes, and then the supernatant was removed. Five volumes of room temperature TRIzol® LS (Invitrogen, Carlsbad, CA, USA) were added and mixed thoroughly. The samples were incubated at 37°C for 5 minutes to ensure the complete deproteinization of nucleic acids. On ice, 1 mL of chloroform was added to the samples, and then the samples were centrifuged at 12000 × g for 30 minutes at 4°C. The upper aqueous layer was transferred to a new tube, and 0.8 volume of pre-chilled isopropanol was added to precipitate the RNA. The tubes were centrifuged at 12000 × g for 30 minutes at 4°C. The supernatant was carefully aspirated off. The pellet was air-dried on ice for 5 minutes, and 30-100 μL of RNase-free non-DEPC-treated water was added. The isolated RNA was quantified using a NanoDrop ND-1000 UV/Vis spectrophotometer, and RNA quality was determined using a Bioanalyzer apparatus (Agilent 2100 Bioanalyzer). Total RNA (6000 ng) from each sample was stabilized in RNAStable (Biomatrica) and sent to Scripps Research Institute for HiSeq Illumina sequencing. For each sample, approximately 0.5 μg of total RNA was depleted of ribosomal RNA using RiboMinus (Life Technologies). Ribo-depleted RNA was prepared for sequencing using an NEBNext Ultra RNA Library Prep Kit (New England Biolabs) for Illumina following the manufacturer’s recommended protocols with 15 PCR cycles. Final libraries were size selected on 2% agarose gels to obtain products with library sizes between 280-380 bases in length. The prepared libraries were loaded onto an Illumina HiSeq 2000 platform for single-end 100 base reads with 7 bases of the index read. The data were processed to generate Fastq files using CASAVA 1.8 and multiplexed based on index sequences.
RNA-Seq data analysis
The reference genome and transcript information for P. falciparum 3D7 v9.3 were obtained from PlasmoDB (http://plasmodb.org/). First, FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to analyze the quality of the reads. Next, reads were mapped to the reference genome using Tophat2 [70] (parameters: -no-coverage-search and -p 24). Samtools was used to select only those reads with a minimum mapping quality of 20 (-q 20). The gene expression profile for each sample was obtained using HTSeq-Count [71] with the parameter “intersection-strict mode”. The total read count for each samples was used in the subsequent analyses. Differential expression analyses following MEL treatment were performed using EdgeR [72]. A log-fold change greater than 1.5 or lower than -1.5, CPM ≥ 1, FDR < 0.05, and p-value < 0.05 were selected as filters for defining the sets of differentially expressed (up- or down-regulated) genes. Differential expression analyses following cAMP treatment were performed using EdgeR [72] and DeSeq [73]. A log-fold change greater than 2.0 or lower than -2.0, CPM ≥ 1, and p-value < 0.05 were selected as filters for defining the sets of differentially expressed (up- or down-regulated) genes.
Functional enrichment analysis
DAVID Bioinformatics Resources v6.7 was used to perform functional enrichment analysis [30, 74]. The web-accessible DAVID program provides a comprehensive set of functional annotation tools to understand the biological meaning behind large lists of genes. The functional categorization is considered significant when the p-value is less than 0.05. To gain more information regarding our set of up- and down-regulated genes, GO terms were assigned manually to those genes using PlasmoDB annotation as a reference. KEGG pathway mapping is used to map molecular datasets, especially large-scale datasets in genomics, transcriptomics, proteomics, and metabolomics, to the KEGG pathway maps for biological interpretation of higher-level systemic functions. Therefore, we used KEGG Mapper (http://www.genome.jp/kegg/tool/map_pathway1.html) against the P. falciparum 3D7 database to identify those pathways that were modulated by cAMP treatment.
ACKNOWLEDGMENTS
This manuscript was corrected by the professional language editing service American Journal Experts (AJE).
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
FUNDING
This work was supported by a grant from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) to CRSG (Process 2011/51295-5) and to DM (Process 2011/50761-2). WRL and GTA received fellowships from FAPESP. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.



- 1. Pineal melatonin: cell biology of its synthesis and of its physiological interactions. Endocr Rev. 1991; 12:151180. [PubMed]
- 2. Melatonin triggers PKA activation in the rodent malaria parasite Plasmodium chabaudi. J Pineal Res. 2011; 50:64-70. [PubMed]
- 3. Calcium-dependent modulation by melatonin of the circadian rhythm in malarial parasites. Nat Cell Biol. 2000; 2:466-468. [PubMed]
- 4. Melatonin and IP3-induced Ca2+ release from intracellular stores in the malaria parasite Plasmodium falciparum within infected red blood cells. J Biol Chem. 2011; 286:5905-5912. [PubMed] https://doi.org/10.1074/jbc.M110.188474.
- 5. Cyclic AMP and calcium interplay as second messengers in melatonin-dependent regulation of Plasmodium falciparum cell cycle. The Journal of cell biology. 2005; 170:551-557. [PubMed] https://doi.org/10.1083/jcb.200505117.
- 6. Ubiquitin proteasome system and the atypical kinase PfPK7 are involved in melatonin signaling in Plasmodium falciparum. J Pineal Res. 2012; 53:147-153. [PubMed] https://doi.org/10.1111/j.1600-079X.2012.00981.x.
- 7. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002; 419:498-511. [PubMed] https://doi.org/10.1038/nature01097.
- 8. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Molecular and biochemical parasitology. 2005; 143:67-79. [PubMed]
- 9. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003; 1:E5. [PubMed] https://doi.org/10.1371/journal.pbio.0000005.
- 10. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003; 301:1503-1508. [PubMed]
- 11. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS pathogens. 2010; 6:e1001223. [PubMed] https://doi.org/10.1371/journal.ppat.1001223.
- 12. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC genomics. 2011; 12:587. [PubMed] https://doi.org/10.1186/1471-2164-12-587.
- 13. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Molecular microbiology. 2010; 76:12-24. [PubMed] https://doi.org/10.1111/j.1365-2958.2009.07026.x.
- 14. RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic acids research. 2011; 39:3820-3835. [PubMed] https://doi.org/10.1093/nar/gkq1223.
- 15. Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC genomics. 2014; 15:150. [PubMed] https://doi.org/10.1186/1471-2164-15-150.
- 16. Estimating mRNA lengths from Plasmodium falciparum genes by Virtual Northern RNA-seq analysis. Int J Parasitol. 2016; 46:7-12. [PubMed]
- 17. Molecular machinery of signal transduction and cell cycle regulation in Plasmodium. Molecular and biochemical parasitology. 2009; 165:1-7. [PubMed] https://doi.org/10.1016/j.molbiopara.2009.01.003.
- 18. Calcium homeostasis in intraerythrocytic malaria parasites. Eur J Cell Biol. 1996; 71:409-413. [PubMed]
- 19. FRET peptides reveal differential proteolytic activation in intraerythrocytic stages of the malaria parasites Plasmodium berghei and Plasmodium yoelii. Int J Parasitol. 2011; 41:363-372. [PubMed]
- 20. Purinergic signalling is involved in the malaria parasite Plasmodium falciparum invasion to red blood cells. Purinergic Signal. 2010; 6:365-372. [PubMed] https://doi.org/10.1007/s11302-010-9202-y.
- 21. The PfNF-YB transcription factor is a downstream target of melatonin and cAMP signalling in the human malaria parasite Plasmodium falciparum. J Pineal Res. 2013; 54:145-153. [PubMed]
- 22. The central role of cAMP in regulating Plasmodium falciparum merozoite invasion of human erythrocytes. PLoS pathogens. 2014; 10:e1004520. [PubMed] https://doi.org/10.1371/journal.ppat.1004520.
- 23. Melatonin signaling and its modulation of PfNF-YB transcription factor expression in Plasmodium falciparum. International journal of molecular sciences. 2013; 14:13704-13718. [PubMed] https://doi.org/10.3390/ijms140713704.
- 24. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS pathogens. 2010; 6:e1000737. [PubMed] https://doi.org/10.1371/journal.ppat.1000737.
- 25. Epigenetics in Plasmodium: what do we really know? Eukaryotic cell. 2010; 9:11501158.. 2010; 6:e1000737. [PubMed] https://doi.org/10.1128/EC.00093-10.
- 26. Antigenic variation in Plasmodium falciparum. Annual review of microbiology. 2008; 62:445-470. [PubMed]
- 27. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryotic cell. 2011; 10:1257-1263. [PubMed] https://doi.org/10.1128/EC.05099-11.
- 28. Tools for accessing and analyzing finished and unfinished sequence data. The Plasmodium Genome Database Collaborative. Nucleic acids research. 2001; 29:66-69. [PubMed] https://doi.org/10.1093/nar/29.1.66.
- 29. An interplay between 2 signaling pathways: melatonin-cAMP and IP3-Ca2+ signaling pathways control intraerythrocytic development of the malaria parasite Plasmodium falciparum. Biochem Biophys Res Commun. 2014; 446:125-131. [PubMed]
- 30. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44-57. [PubMed]
- 31. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009; 37:1-13. [PubMed] https://doi.org/10.1093/nar/gkn923.
- 32. Plasmodium kinases as targets for new-generation antimalarials. Future Med Chem. 2012; 4:2295-2310. [PubMed]
- 33. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC genomics. 2005; 6:30. [PubMed] https://doi.org/10.1186/1471-2164-6-30.
- 34. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS pathogens. 2005; 1:e44. [PubMed] https://doi.org/10.1371/journal.ppat.0010044.
- 35. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC genomics. 2004; 5:79. [PubMed] https://doi.org/10.1186/1471-2164-5-79.
- 36. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Molecular microbiology. 2007; 63:391-403. [PubMed]
- 37. Substrate specificity and nucleotides binding properties of NM23H2/nucleoside diphosphate kinase homolog from Plasmodium falciparum. J Bioenerg Biomembr. 2010; 42:361-369. [PubMed]
- 38. Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase. J Virol. 2009; 83:7142-7150. [PubMed] https://doi.org/10.1128/JVI.00511-09.
- 39. Primary structure and sexual stage-specific expression of a LAMMER protein kinase of Plasmodium falciparum. Int J Parasitol. 2001; 31:387-392. [PubMed]
- 40. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol Cell Biol. 2003; 23:4139-4149. [PubMed] https://doi.org/10.1128/MCB.23.12.4139-4149.2003.
- 41. Phylogenomic analysis and evolution of the potassium channel gene family. Receptors Channels. 2003; 9:363-377. [PubMed]
- 42. Melatonin feedback on clock genes: a theory involving the proteasome. J Pineal Res. 2015; 58:1-11. [PubMed]
- 43. Substrate targeting in the ubiquitin system. Cell. 1999; 97:427-430. [PubMed]
- 44. Melatonin-induced temporal up-regulation of gene expression related to ubiquitin/proteasome system (UPS) in the human malaria parasite Plasmodium falciparum. International journal of molecular sciences. 2014; 15:22320-22330. [PubMed] https://doi.org/10.3390/ijms151222320.
- 45. Melatonin and ubiquitin: what’s the connection? Cellular and molecular life sciences :: CMLS. 2014; 71:3409-3418. [PubMed]
- 46. Polyamines. Annual review of biochemistry. 1984; 53:749-790. [PubMed]
- 47. The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 A resolution reveals a novel fold. Structure. 1999; 7:583-595. [PubMed]
- 48. Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 1999; 7:567-581. [PubMed]
- 49. In the human malaria parasite Plasmodium falciparum, polyamines are synthesized by a bifunctional ornithine decarboxylase, S-adenosylmethionine decarboxylase. J Biol Chem. 2000; 275:8097-8102. [PubMed]
- 50. Parasite-specific inserts in the bifunctional S-adenosylmethionine decarboxylase/ornithine decarboxylase of Plasmodium falciparum modulate catalytic activities and domain interactions. The Biochemical journal. 2004; 377:439-448. [PubMed] https://doi.org/10.1042/BJ20030614.
- 51. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome biology. 2003; 4:R64. [PubMed] https://doi.org/10.1186/gb-2003-4-10-r64.
- 52. Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure. 2005; 13:963-971. [PubMed]
- 53. (2014). Transport of mRNA Through the Nuclear Pore in Plasmodium falciparum. In: Hommel M and Kremsner PG, eds. Encyclopedia of Malaria: Springer New York), pp. 1-8. [PubMed]
- 54. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to aminoacid starvation. Malaria journal. 2009; 8:99. [PubMed] https://doi.org/10.1186/1475-2875-8-99.
- 55. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS pathogens. 2009; 5:e1000569. [PubMed] https://doi.org/10.1371/journal.ppat.1000569.
- 56. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic acids research. 2009; 37:2596-2606. [PubMed] https://doi.org/10.1093/nar/gkp115.
- 57. Plasmodium falciparum signal peptidase is regulated by phosphorylation and required for intra-erythrocytic growth. Molecular and biochemical parasitology. 2008; 157:137-147. [PubMed]
- 58. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010; 328:910-912. [PubMed] https://doi.org/10.1126/science.1188191.
- 59. Characterization of a PRL protein tyrosine phosphatase from Plasmodium falciparum. Molecular and biochemical parasitology. 2008; 158:1-10. [PubMed]
- 60. Antibodies to PfSEA-1 block parasite egress from RBCs and protect against malaria infection. Science. 2014; 344:871-877. [PubMed] https://doi.org/10.1126/science.1254417.
- 61. Co-inhibition of Plasmodium falciparum S-adenosylmethionine decarboxylase/ornithine decarboxylase reveals perturbation-specific compensatory mechanisms by transcriptome, proteome, and metabolome analyses. J Biol Chem. 2009; 284:4635-4646. [PubMed] https://doi.org/10.1074/jbc.M807085200.
- 62. Protein kinases of malaria parasites: an update. Trends Parasitol. 2008; 24:570-577. [PubMed]
- 63. Plasmodium falciparum possesses a cell cycle-regulated short type replication protein A large subunit encoded by an unusual transcript. J Biol Chem. 2002; 277:17493-17501. [PubMed]
- 64. Centrins, cell cycle regulation proteins in human malaria parasite Plasmodium falciparum. J Biol Chem. 2008; 283:31871-31883. [PubMed]
- 65. One billion years of Myb. Oncogene. 1996; 13:223-235. [PubMed]
- 66. Human malaria parasites in continuous culture. Science. 1976; 193:673-675. [PubMed]
- 67. Synchronization of Plasmodium falciparum erythrocytic stages in culture. The Journal of parasitology. 1979; 65:418-420. [PubMed]
- 68. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum. Molecular microbiology. 2005; 55:184-196. [PubMed]
- 69. Strand-specific RNA-seq applied to malaria samples. Methods Mol Biol. 2012; 883:59-73. [PubMed] https://doi.org/10.1007/978-1-61779-839-9_4.
- 70. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013; 14:R36. [PubMed] https://doi.org/10.1186/gb-2013-14-4-r36.
- 71. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166-169. [PubMed] https://doi.org/10.1093/bioinformatics/btu638.
- 72. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139-140. [PubMed] https://doi.org/10.1093/bioinformatics/btp616.
- 73. Differential expression analysis for sequence count data. Genome biology. 2010; 11:R106. [PubMed] https://doi.org/10.1186/gb-2010-11-10-r106.
- 74. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009; 37:1-13. [PubMed] https://doi.org/10.1093/nar/gkn923.
Last Modified: 2016-10-03 21:18:39 EDT

