Genes & Cancer
Genetic determinants in head and neck squamous cell carcinoma and their influence on global personalized medicine
Nicole L. Michmerhuizen1,2, Andrew C. Birkeland1, Carol R. Bradford1,3, J. Chad Brenner1,3
1 Department of Otolaryngology – Head and Neck Surgery, University of Michigan Medical School, Ann Arbor, MI, USA
2 Department of Pharmacology, University of Michigan Medical School, Ann Arbor, MI, USA
3 Comprehensive Cancer Center, University of Michigan Medical School, Ann Arbor, MI, USA
Correspondence to: J. Chad Brenner, email: [email protected]
Keywords: epidemiology, head and neck squamous cell carcinoma, human papillomavirus, personalized medicine, sequencing
Received: May 05, 2016
Accepted: July 21, 2016
Published: July 28, 2016
ABSTRACT
While sequencing studies have provided an improved understanding of the genetic landscape of head and neck squamous cell carcinomas (HNSCC), there remains a significant lack of genetic data derived from non-Caucasian cohorts. Additionally, there is wide variation in HNSCC incidence and mortality worldwide both between and within various geographic regions. These epidemiologic differences are in part accounted for by varying exposure to environmental risk factors such as tobacco, alcohol, high risk human papilloma viruses and betel quid. However, inherent genetic factors may also play an important role in this variability. As limited sequencing data is available for many populations, the involvement of unique genetic factors in HNSCC pathogenesis from epidemiologically diverse groups is unknown. Here, we review current knowledge about the epidemiologic, environmental, and genetic variation in HNSCC cohorts globally and discuss future studies necessary to further our understanding of these differences. Long-term, a more complete understanding of the genetic drivers found in diverse HNSCC cohorts may help the development of personalized medicine protocols for patients with rare or complex genetic events.
INTRODUCTION
Recent next generation sequencing (NGS) studies of head and neck squamous cell carcinomas (HNSCC) have shed light onto the underlying genetic profiles for this aggressive disease [1, 2] and enabled a move towards personalized medicine, in which therapy is guided by tumor genetics. Notably, however, the vast majority of patients sequenced thus far have been restricted to a single epidemiologic population—human papillomavirus (HPV) negative, Caucasian, and high tobacco and/or alcohol use. There has been little information on the genetic profiles in other epidemiologic cohorts; thus, the genomic events driving pathogenesis in these patients remain poorly understood. The rationale to overcome this void is clear and detailed below.
In the US and other high-income countries, personalized medicine approaches are increasingly being applied for many advanced cancers including HNSCC [2-4]. Personalized medicine protocols, such as the National Cancer Institute-Molecular Analysis for Therapy Choice (NCI-MATCH) trial, seek to test molecularly targeted therapies in patients with corresponding mutations [5-7]. However, these protocols often rely on targeted NGS approaches, which are resource intensive and unlikely to be implemented in low- or middle-income countries in the near future. Thus, the idea of targeted and personalized therapy may need to be adjusted in areas where sequencing-based medicine is not yet achievable. One way to do this is to understand the genetic events common to different epidemiologic populations and guide biomarker-based research and medicine towards the most frequent and tractable biomarkers in the region.
Genetic studies comparing ethnic and epidemiologic sub-groups have also been very informative in generally understanding oncogenes and tumor suppressors in cancer. As an example of differential distributions of genetic events based on ethnicity, TMPRSS2:ETS gene fusions are found in approximately 50% of prostate cancers in the US, but only 10% of prostate cancers in China. As a result, focused deep sequencing of TMPRSS2:ETS gene fusion negative Chinese prostate cancers identified high frequency and previously unrecognized genomic events in alternative pathways [8, 9]. Similarly, we recently performed NGS analysis of an epidemiologically low risk HNSCC (from a young, non-smoker/drinker, HPV-negative patient) with the hypothesis that the tumor would have relatively few mutations compared to a tobacco-related HNSCC. Indeed, our analysis found a potential driver amplification of the tyrosine kinase receptor FGFR1. Extending the discovery to The Cancer Genome Atlas (TCGA) HNSCC cohort, we demonstrated that the FGF/FGFR pathway is dysregulated in >30% of HNSCCs and likely represents a previously unrecognized aberration driving disease pathogenesis [10]. Consequently, carefully designed studies focusing on the genetics of under-studied epidemiologic populations can be very informative.
In this review, we will discuss current knowledge of the variations in prevalence, environmental factors, and genetic factors in HNSCC across different regions from around the world. All large (50 or more total patients), available NGS studies in head and neck cancer were included along with other sequencing studies, which were identified primarily using the most recently published PubMed articles after searching for relevant terms. This review also includes discussion of the variation in HNSCC incidence and severity evidenced in black and white American cohorts [11-14]. (In this review, we will use the New England Journal of Medicine convention of black as opposed to African American [15]). It is evident from these early studies that different epidemiologic subsets of HNSCC may associate with different tumor genetics and unique outcomes, and thus may be responsive to different targeted therapies.
HNSCC Rates Globally
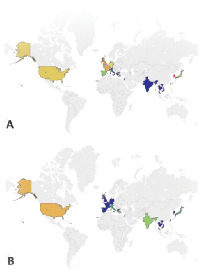
Historically, different rates of HNSCC have been evidenced in different epidemiologic populations (Figure 1, Table S1). While environmental factors are thought to be a major contributor to this variability, it is unclear if the underlying acquired genetic events are similar across cohorts. Furthermore, the mutational effects of other factors associated with HNSCC globally (most notably high risk HPV strains 16 and 18, but also betel nut in Southeast Asia, nitrosamines in Asia, Epstein-Barr virus (EBV) in Africa and Asia) have been examined in some studies but still require further characterization [16-18]. Here, we will review what is known about HNSCC incidence and mortality in representative countries from around the world.
Developed Countries: United States, Canada, and Europe
Two thirds of HNSCC cases occur in developed countries, where the use of tobacco and alcohol is prevalent [19]. Odds ratios for developing HNSCC due to tobacco and/or alcohol use are 3-4 times higher in Europe and Latin America, where the use of both substances is more widespread, than in North America [20]. In general, between 1983 and 2002, incidence rates for oral cavity cancers (for which increased risk is particularly noted in smokers) increased in Europe and decreased in the US and Canada [21]. During this time period, incidence of oropharyngeal cancer also increased in eastern and northern Europe. These trends may reflect changes in the proportion of the population using tobacco and/or alcohol.
Tobacco use alone, however, does not account for variation in HNSCC throughout Europe. Based on rates reported by Simard et al., HNSCC incidence in all anatomic subsites is somewhat increased in France compared to eastern European countries and is markedly higher in France compared to other European nations (such as the UK and Italy) [21]. Of the representative European countries in Figure 1, however, smoking rates are similar in France, Italy, and the UK. Increased tobacco use, then, cannot completely explain the increased rate of oral cavity cancers between France and other European nations or the higher incidence of cancers of other sites in men from Italy as compared to the UK [21]. While HNSCC in France may be driven somewhat by elevated levels of tobacco use, other factors, including biological differences, may also be crucial for increased prevalence.
Asia
Increased rates of HNSCC, particularly oral cavity squamous cell carcinoma, in southern and southeastern Asian countries are often attributed to betel quid exposure [22]. Head and neck tumors are one of the most common malignancies in males in some parts of south central Asia. Parkin et al. identified the highest incidence of oral cancer in Melanesia (31.5 per 100,000 in men, 21.2 per 100,000 in women) [23]. While nasopharyngeal tumors also have greatest incidence in southeastern Asia, trends in oropharynx cancer vary by specific country [23]. Fewer oral cancer cases are observed in Chinese and Middle Eastern cohorts, where betel quid is used more rarely, as compared to other Asian countries [24]. High rates of laryngeal and other types of HNSCC in China may be due in part to increased tobacco use in this country. Lower incidences of HNSCC at all sites in the Middle East are possible for a variety of reasons, including, but not limited to, the lower use of betel quid, tobacco, and alcohol in this region.
Africa
There is relatively little data available on HNSCC in African cohorts; nevertheless, tumor epidemiologic differences may exist. A systematic review of the literature since 1990 by Faggons et al. found that 7750/8861 (87.5%) patients with HNSCC in sub-Saharan Africa presented with cancer of the oral cavity or oropharynx [25]. Subsite specificity may vary significantly between countries; the nasopharynx was the most common site identified in a review of the literature on head and neck cancer in Nigeria [26], but there were much less frequent reports of tumors of the nasopharynx, nasal cavity, or paranasal sinuses (410/8861, 4.8%), larynx (385/8861, 4.5%), or hypopharynx (66/8861, 0.8%) in sub-Saharan Africa [25]. These differences may be due to difficulties in screening for cancers in these subsites. Consistent with prevalence in the sub-Saharan cohort, oral cavity and oropharyngeal tumors accounted for 27/46 (58.7%) cases in black TCGA patients while larynx cancer was also common (18/46, 39.1%) and hypopharynx tumors were infrequent (1/46, 2.2%).
Within Africa, reports of HNSCC incidence vary widely, from 0.8 per 100,000 in Ghana [27] to 11.1 per 100,000 in South Africa [23]. Tumors of the pharynx and larynx are the second and seventh most common types of cancer seen at the Korle Bu teaching hospital in Ghana, representing 7.4% and 3.5% of all malignancies, respectively [28]. Furthermore, age at tumor presentation in African patients was approximately 20 years younger than in American populations, which may be explained by biological, exposure and/or other differences between these populations [25]. Several epidemiologic factors may contribute to differences in African HNSCC including HIV infection, which has been shown to increase the risk of HNSCC by two to three times in the US [29]. Despite this fact, the role of HIV in African HNSCCs is unclear due to a lack of studies comparing HIV positive and negative patients; anecdotal evidence suggests that HIV positive patients have poorer clinical outcomes but further comparison is necessary [25].
Variation within Geographic Regions
Variation in HNSCC rates is also observed within different ethnic groups within specific geographic regions [30]. For example, Ho et al. considered HNSCC incidence and mortality rates in three Taiwanese tribal groups (Fukkien, Hakkas, and Aboriginal) [31]. Between 1979 and 1997, compared to Fukkien groups, HNSCC mortality rates decreased in Hakka and increased in Aboriginal tribes. Incidence trends between 1979 and 1996 were similar, particularly in Aboriginals with high chewing prevalence for betel quid. While environmental and socioeconomic factors vary between groups, these differences alone may not explain the observed variation in incidence of and mortality from HNSCC. Interestingly, genetic differences have been noted between Fukkiens and Aboriginals in alleles responsible for metabolic activation of carcinogenic nitrosamines [32].
Additionally, significant differences in HNSCC incidence, particularly for larynx cancer, have been noted between black and white Americans. In a study by DeSantis et al., there was a higher incidence of laryngeal cancer in black (10.4/100,000) compared to white males (6.6/100,000); differences were not noted in the incidence of tumors at other HNSCC subsites [11]. Goodwin et al. evidenced 15% and 77% increased incidence of oral cavity/pharynx and larynx tumors, respectively, in black as compared to white male Americans. These authors also observed increased incidence of larynx cancer (but not other sites) in black females [12]. The differences in this study were observed to the greatest degree in patients under the age of 65, suggesting that hereditary or early onset factors may be involved. Furthermore, black women displayed higher rates of non-oral cavity, non-oropharyngeal HNSCCs compared to white and other ethnic female cohorts worldwide [21].
HNSCC incidence varies not only by subsite but also by severity and survival rates in groups of black and white American patients. Black patients are more likely to present with late stage, poor prognosis HNSCC than whites [12, 13]. Regardless of cancer site, mortality for black males is on the order of two times higher when compared to that of similar white patients [12]. Furthermore, independent of cancer stage, compared to 63% of whites, only 42% of blacks survive five years following HNSCC diagnosis [11]. While socioeconomic and environmental factors certainly may play a role in this survival difference, the fact that these differences are seen in younger patients suggests a potential role for genetic factors in tumor aggressiveness [14]. Further investigation is necessary to determine these potential hereditary factors and how they may diverge between ethnic cohorts to cause more aggressive disease phenotypes.
High Risk Human Papillomavirus is Changing HNSCC Epidemiology
Infection with high risk HPV is a major risk factor for head and neck cancers, particularly in the oropharynx [33-40]. Gillison et al. reported that HPV positive HNSCCs occurred in 34/60 (56.7%) patients with oropharyngeal cancer but only 10/84 (11.9%) oral cavity, 16/86 (18.6%) larynx, 2/21 (9.5%) hypopharynx, and 0/2 (0%) nasopharynx tumors [36]. Largely due to the increased incidence of the oropharyngeal HNSCC subtype, the recent HPV epidemic is contributing to a rapid change in the epidemiologic distribution of HNSCC globally. To place this in perspective, HPV-positive oropharyngeal squamous cell carcinoma incidence surpassed that of invasive cervical cancer in 2013 [41]. While cervical lesions are often diagnosed early and treated pre-neoplastically to avoid disease progression [42, 43], most oropharyngeal cancer patients present with advanced stage III/IV disease [39]. 70-80% of HPV-positive oropharynx cancers respond to intensive therapy consisting of chemoradiation or surgery in most series [44, 45]. The remaining 20-30% of patients’ tumors progress to lethal recurrent or metastatic disease, indicating the need to define biomarkers that will predict the subset of patients that would benefit from more aggressive therapy and will minimize morbidity in less difficult cases. Studies indicating improved prognosis for HPV-positive patients have also suggested that de-escalated chemoradiation therapies, which reduce toxicity and improve quality of life, may be more effective in oropharyngeal, HPV-positive than HPV-negative HNSCCs [46-49]. However, this is not the case for other HNSCC subsites, where rates of HPV infection are typically quite low and might not be an established etiologic factor. Consequently, there are no paradigms for the treatment of oral cavity, laryngeal, hypopharyngeal, or nasopharygeal cancers; additionally, the effects of HPV status on outcomes assessment in these other subsites remains underexplored [50].
In the face of the HPV epidemic, high risk viral infection has been associated with oropharyngeal cancers in studies from across the world. A systematic review by Stein et al. compared the prevalence of HPV-positive oropharynx cancer in 23 countries worldwide [51]. Taiwan, Canada, and the Czech Republic had the highest prevalence of HPV-positive oropharyngeal cancer, with much lower HPV burdens in the Netherlands, Brazil, and Spain. Overall, the results of this analysis suggest that HPV-positive tumors may be more common in developed countries. In another systematic review, Mehanna et al. identified HPV-positive cancers of the oropharynx in 59.9% of 2550 North American patients, but only 39.7% of 2278 European patients and 32.5% of 568 patients from other regions [52]. An additional cohort of 31/67 (46.2%) Australian patients also displayed high prevalence of HPV-positive oropharyngeal cancer [53]. Chinese patients displayed lower infection rates with 43/207 (20.8%) HPV-positive oropharyngeal tumors and 36/124 (29.4%) HPV-positive tonsil lesions [54]. The prevalence of HPV-positive oropharyngeal cancer was also lower across Africa as compared to many developed countries. 5/125 (4%) South African men were identified with HPV-positive oropharyngeal cancer, and only two of these cases were high risk HPV [55]. Similarly, 0/22 (0%) oropharyngeal cancer and 2/29 (6.9%) oral tongue cancer patients in Mozambique tested positive for HPV infection [56]. In Senegal, only 4/117 (3.4%) HNSCC patients had HPV-related tumors. Of the five patients in this study with oropharyngeal cancer, none tested HPV positive [57].
Of note, the use of numerous testing methodologies to assess a diverse array of HPV variants may introduce inconsistency in HPV detection outcomes. HPV16 accounts for over 90% of HPV-positive HNSCC cases [58] and can be detected by assessing HPV DNA, HPV RNA, viral oncoprotein, cellular protein and/or HPV-specific serum antibody levels [59]. Staining for p16 by immunohistochemistry has historically been used as a surrogate for HPV status, but does not assess for actual HPV infection. Reverse transcriptase (RT)-PCR, PCR-mass spectrometry and in situ hybridization (ISH)-based protocols are also used to detect HPV-positive HNSCC in some cases [60, 61]. Thus, consideration of further studies with controlled testing of the global prevalence of HPV-positive oropharyngeal cancer will be important, particularly in Africa given the discordance between high cervical and low oropharyngeal HPV-positive cancer rates in this region.
While rates of HPV infection worldwide are not fully realized, evidence does indicate that prevalence may be rising globally and may drive increased incidence of HNSCC. Considering the large proportion of the population that is infected with HPV [62], malignant transformation is comparatively rare as HPV infections are usually cleared quickly [63]. In rare cases, however, genomic instability and unrestricted proliferation caused by viral oncogene activity lead to tumorigenesis. Cervical infection with HPV, if not cleared, can lead to precancers in the genital area as well as the head and neck region through sexual contact. Thus, as a high-level surrogate for oropharyngeal cancer prevalence, we can analyze the reports of HPV prevalence in women, noting a wide range of cervical infection rates in cohorts worldwide [64-66]. Based on a meta-analysis of women with normal cytology, Bruni et al. estimated that the regionally-adjusted prevalence of high risk HPV infection, as detected by polymerase chain reaction (PCR) or Hybrid Capture 2 (a DNA hybridization assay for detecting HPV strains with a fluorescent readout), in females is 47,271/851,901 (5.0%) worldwide. The prevalence of both low and high-risk HPV strains is 73,019/1,016,719 (11.7%), which varies between rates as high as 75/225 (35.4%) in the Caribbean to those as low as 31/1,435 (1.7%) in Western Asia [66]. Of all viral strains, HPV16 was the most commonly detected in this study but tended to correlate inversely with overall HPV prevalence. In the more limited analysis of data from male patients, an important population given the increased prevalence of HNSCC in this group, a similar overall prevalence (182/1139, 16%) and even more significant amount of variation was observed, particularly when separating low- and high-risk groups [67, 68]. We compared adjusted HPV infection prevalence with oropharyngeal cancer incidence in cohorts worldwide to consider possible associations between these two variables (Figure 2). In a subset of countries, both increased HPV prevalence and oropharyngeal cancer incidence were observed. (For example, women in Eastern Europe have high prevalence of cervical HPV infection (904/4053, 21.4%), and the oropharyngeal cancer incidence in Slovakian men is also elevated (15.4/100,000)).Alternatively, in other regions, decreased HPV prevalence and increased oropharyngeal cancer incidence were noted. (In India, only 1816/23,061 (7.1%) women tested positive for HPV infection, but 9.1/100,000 men develop oropharyngeal tumors [66, 69]). These findings may be explained by regional differences in HPV infection rates; however, developing a more complete understanding of this relationship is additional motivation for controlled HPV testing in global cohorts.
Sexual activity, particularly oral sex with multiple partners, increases HPV-positive cancer risk at all sites. As is the case with other risk factors, sexual practices are widely divergent across populations globally and may be used to at least partially explain differences in the incidence of HPV-positive HNSCCs in various populations. For instance, 78% of American, but only 9% of Indian men reported ever having oral sex, and individuals born after 1960 have more commonly engaged in this activity [70]. Men are also more likely to have multiple partners than women [71-73] and the prevalence of HPV infection was much higher in high-risk populations of males as compared to females with similar numbers of sexual partners [74]. Given these differences, it stands to reason that HPV-positive HNSCC rates will vary widely between cohorts globally.
Variation between HPV-positive head and neck cancer rates has already been observed in whites and blacks in the US and may atleast partially explain racial disparity in survival rates. Settle et al. found that differences in the median overall survival (OS) of an American cohort were driven primarily by differences in tumors of the oropharynx (white 69.4 months, black 25.2 months, p = 0.0006) and not by tumors at other sites (white 17.1 months, black 17.5 months, difference n.s.) [75]. In a separate analysis of OS for black and white Americans with HPV-positive or HPV-negative HNSCC, Jiron et al. determined that the hazard ratio was greatest for black patients with tumors of the oropharynx but that adjustment for HPV status drastically reduced this ratio to a value close to unity [76]. These results suggest that the poorer prognosis of black patients in the US may be due to the reduced rate of HPV-positive oropharyngeal cancers in this group. Nevertheless, this does not explain differences in laryngeal cancer incidence and survival statistics, and further studies are necessary to more fully evaluate this hypothesis.
Variation in Genetic Landscape between Epidemiologic Sub-Groups
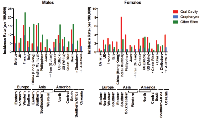
Despite the majority of HNSCCs occurring in non-American populations (incidence rate of 60,000 annually in the US vs 490,000 annually in the rest of world), NGS studies have been limited to cohorts of primarily European ancestry and not other ethnic groups or epidemiologic populations. TCGA and the International Cancer Genome Consortium (ICGC) reported sequencing for cohorts of HNSCC patients in the United States and India, respectively [1, 18]. In the TCGA cohort, the majority of these patients were white (242/279, 86.7%), with only 25/279 (9.0%) black. Different mutational profiles were evidenced between black and white HPV-negative patients (Figure 3). For instance, black patients have significantly higher rates of BIRC2/3 amplification compared to white HNSCC patients in this study (25.0% vs 4.3%, p < 0.001). Although they do not reach statistical significance, blacks also trend toward decreased EGFR (4.2% vs 13.5%, p = 0.19) and increased FGFR1 (16.7% vs 9.1%, p = 0.24) amplification. Other genetic aberrations were similar between these two ethnic groups [77, 78].
ICGC data also noted distinct differences in genomic aberrations in Indian patients with oral cancer of the gingivo-buccal region [18]. When compared to white American HNSCC TCGA patients, Indian patients display significantly lower rates of EGFR (p = 0.0365) and MYC (p = 0.0365) amplification, PIK3CA activation by mutation or amplification (p = 0.0027) and CDKN2A deletion (p = 0.0045) (Figure 3). As identified in previous studies of HNSCC, the authors observed frequent copy number alterations or mutations in TP53, FAT1, CASP8, HRAS and NOTCH1 in the Indian HNSCC patients [79]. Interestingly, the ICGC study also identified five genes (USP9X, MLL4, ARID2, UNC13C and TRPM3) and three pathways (Wnt signaling, dorso-ventral axis formation and axon guidance) previously not associated with HNSCC in TCGA. These events may be specific to this epidemiologic sub-group. For example, mutations in TRPM3 were identified in 5/50 (10%) Indian ICGC patients, but only 4/208 (1.9%) white and 1/25 (4%) black HPV-negative TCGA patients. USP9X was more frequently mutated or deleted in Indian (7/37, 18.9%) than in white (23/208, 11.1%) or black (1/25, 4%) HPV-negative patients [77, 78]although this difference did not reach significance using the Chi-square test with Yate’s correction (p = 0.2837). Furthermore, the frequency of copy number alteration or mutation of FAT1, FAT3, and FAT4 were increased in the Indian ICGC cohort [18].
While there have been few other in-depth genomic studies of large ethnic or epidemiologic cohorts, unique mutational profiles are likely to exist in other global populations. Vettore et al. recently performed targeted deep sequencing on a cohort of 60 patients treated in Singapore and found that mutation frequencies for TP53, CDKN2A, and NOTCH1 were infrequent compared to other studies. While TP53 and CDKN2A represent the first and third most commonly mutated genes in the overall TCGA HNSCC cohort, mutations in these genes, respectively, were present in only 23/60 (38.3%) and 3/60 (5%) of Singaporean patients. Furthermore, NOTCH1 mutation was identified in significantly fewer of these patients (3/60, 5%) than white HPV-negative TCGA patients (41/208, 14.6%) (p = 0.012). Conversely, DST, RNF213, COL6A6 and ZFHSX4 mutations were observed much more commonly in patients in the Singaporean study. Similar studies that could reveal additional trends in other epidemiologic populations, therefore, are clearly warranted.
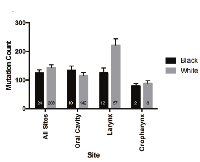
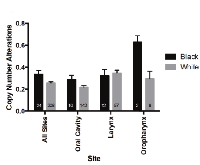
The mutational loads were similar between white and black cohorts in TCGA (Figure 4) and Indian patients in ICGC data (mean total number of mutations: 112.79 +/- 19.25) [18]. Copy number alterations, however, were increased in black as compared to white patients (0.3342 vs 0.2560, p = 0.0443) (Figure 5). When subsites were considered individually, trends between ethnic cohorts were apparent for copy number alterations in oropharyngeal tumors (however only 2 black patients compared to 8 white) and were also observed for tumors of the oral cavity (0.2884 vs 0.2174, p = 0.17). These comparisons are limited by the small number of black patients included in this analysis and may be due to sample bias.
Worldwide rates of established HNSCC molecular events
Currently, EGFR, PIK3CA, NOTCH pathway and TP53 genes are among the most frequently altered in HNSCC. As these genes have long been associated with HNSCC pathogenesis, several smaller cohort studies have been published assessing the rates of genomic events for these genes in various ethnic and epidemiologic populations. Understanding which populations have unique genetic landscapes may aid in selecting the most informative populations for immediate NGS analysis. Unfortunately, a relatively small number of studies have sequenced HPV-positive HNSCCs in international analyses. Thus, we will separately address worldwide genomic event frequencies in HPV-negative and HPV-positive disease when possible.
Epidermal Growth Factor Receptor (EGFR)
It has been recognized for nearly 30 years that EGFR is overexpressed in the majority of HNSCC tumors [80]. The effects of EGFR activation in squamous cells may be pleiotropic: not only can changes in receptor signaling affect the Ras-MAPK, PI3K-AKT-PTEN, and/or phospholipase C pathways, but they may also activate other receptors by ligand-independent dimerization. Consistent with the prevalence of EGFR protein overexpression and demonstrating its importance, EGFR-targeted antibody, cetuximab, is currently the only FDA-approved targeted therapy for HNSCC and has been shown to improve overall survival of patients [81, 82]. A meta-analysis of 37 studies by Keren et al. examined EGFR levels in surgically resected primary tumor samples and identified 1948/3346 (57.8%) cases with high protein expression. The majority of patients were from Europe, with some cohorts also from the US and east Asia; overexpression was frequently and consistently noted in Austrian, Spanish, and Dutch cohorts, while it was infrequent in Swedish, French, and Italian populations [83]. High levels of EGFR expression may also be more common in Sudan, where 126/150 (84%) head and neck cancer cases displayed overexpression by IHC analysis [84].Despite frequent overexpression of EGFR, rates of genetic aberration (by amplification or mutation) are relatively low (14.3% in TCGA) [1] and a small number of studies have assessed EGFR genomic aberrations in individual populations. For example, 3/41 (7.3%) Korean patients, most of whom had larynx cancer, displayed a mutation in the kinase domain of EGFR [85], but similar mutations were much less commonly observed in Caucasian or Spanish patients [86-88]. One TCGA HNSCC patient displayed truncating EGFRvIII mutation. This particular mutation was previously detected by Sok et al. in 14/33 (42.4%) HNSCCs along with wild type EGFR and was correlated with resistance to targeted EGFR therapy [89]. While mutations of EGFR in particular may be somewhat infrequent, this pathway as a whole is often aberrantly expressed. For example, 31/60 (52%) Singaporean patients with tongue cancer displayed mutations in the EGFR pathway (ADCY8, AKT3, COL1A1, COL1A2, EPN1, FGFR3, FGFR4, HRAS, HSP90AA1, IGF1R, ITGAV, ITGB3, JAK2, JAK3, MTOR, PDGFRA, PIK3CD, PIK3CG, PTEN, RASA1, SIPA1, and PIK3CA), and 46/208 (22.1%) of the TCGA cohort displayed amplification of EGFR family members (EGFR, ERBB2-4, EGF, NRG1-4, EREG, AREG, TFGA, BTC, and HBEGF) [1, 17]. An improved understanding of the genetic and/or biochemical mechanisms driving EGFR overexpression in HNSCC will be necessary before extending the assessment of EGFR mechanisms globally. However, as global cohorts are being prioritized for genetic studies, populations such as Swedish and Italian, which have been associated with lower overall rates of EGFR overexpression, may show the largest variation in genomic landscape to those HNSCCs already sequenced. Although EGFR copy number, mutation status, and expression level have not been correlated with patient response to EGFR targeted therapies to date, future studies might reveal uses for these characteristics as biomarkers; differences between HNSCC cohorts from various geographic regions could stratify patient groups that might be more responsive to anti-EGFR or other combination treatments.
Catalytic Subunit of Phosphotidylinositol 3-kinase (PIK3CA)
The PI3K-AKT-PTEN pathway has been identified as the most frequently mutated or amplified oncogenic pathway in HNSCCs in the TCGA cohort [1, 90]. Mutations and/or amplifications in PIK3CA, the gene encoding the catalytic subunit of phosphotidylinositol 3-kinase (PI3K), are the most common alterations in this pathway and are observed in 36.9% of the TCGA HNSCC cohort [77, 78]. These aberrations lead to increased cell growth and viability, may drive tumor progression, and are more commonly observed in advanced stage disease as reviewed elsewhere [91, 92]. PIK3CA mutation or amplification is also more frequent in HPV-positive tumors, including in the TCGA cohort (56% HPV-positive vs 34% HPV-negative) [1, 90, 93]. Loss of function of PTEN also results in failure to “turn off” PI3K signaling and is observed in an additional 10% of HNSCC patients [94].Rates of PIK3CA aberration have been assessed in various global cohorts [95-100] and are shown in Figure 6. Low frequencies of gene amplification were observed in 3/33 (9%) patients in a German HNSCC cohort as well as 3/115 (2.3%) and 6/50 (12%) individuals in two independent Japanese groups [96, 98, 101]. Alternatively, Redon et al. noted 6/9 (66.6%) French HNSCC patients have increased copy number [102]. PIK3CA mutation is generally less common than gene amplification. Somatic mutations, commonly including “hotspot” amino acid changes to the kinase (H1047R) or helical (E545K, E542K) domains, occur in 20.8% of the TCGA cohort, which is consistent with rates of ~10-20% in other studies [95-97]. PIK3CA mutation rates greater than 10% were noted in cohorts from Thailand (6/58), India (2/19), and Israel (4/37) [96, 97, 100]. Surprisingly, a complete lack of PIK3CA mutations in the helical or kinase domains (exons 9 and 20) were observed in populations of 18 Vietnamese, 33 German, and 86 Greek patients as detected by PCR [97-99]. This may be due to increased activation of HRAS, which signals upstream of PIK3CA, in these epidemiologic sub-groups [103]. Due to the variation in PIK3CA mutation rates between 1/35 (2.9%) and 5/24 (20.8%) in US patient populations [95, 104] and the relatively small number of HNSCC tumor samples that have been sequenced worldwide, additional cohort studies are warranted to further consider potential associations between rates of genetic aberration and patient ethnicity or epidemiologic risk.
NOTCH pathway genes
In 2011, sequencing-based analysis of HNSCC tumors led to the discovery of inactivating NOTCH pathway alterations as the third most common molecular event in the disease [1, 77, 79, 104]. In fact, mutations in one gene from this pathway, NOTCH1, were observed in ~15% of samples in addition to less frequent mutations of the NOTCH2 (~5%) and NOTCH3 (~4%) genes, with rare copy number alterations reported. Soon after these reports were published, and in contrast to the prevalent loss-of-function mutations, copy number increases and overexpression of the NOTCH ligands JAG1 and JAG2 and the receptor NOTCH3 were found in a small subset of predominantly white HNSCCs [105]. Importantly, the functional role of both the activating and inactivating alterations have yet to be fully characterized. In T lymphoplastic leukemia and other hematologic malignancies, the NOTCH pathway has long been known to have an oncogenic function [106], but the role of alterations in NOTCH1 and other related genes in solid tumors is still the topic of extensive research. NOTCH pathway alterations have been reviewed elsewhere [107], but they are suggested to regulate squamous cell differentiation in multiple model systems of HNSCC. Consistent with this notion, many HNSCCs are characterized by recurrent mutations in the TP63, IRF6 or MED1 genes, which have also been suggested to regulate squamous differentiation, supporting a functional importance of this pathway for HNSCC pathogenesis [79]. Rudy et al. also recently showed that inhibition of WNT signaling via a PORCN inhibitor reduced the metastatic spread of numerous patient-derived HNSCC cell lines in an in vivo chicken chorioallantoic membrane model; this suppression occurred with greater efficacy in cell lines with NOTCH1 deficiency [108].
While the discovery of NOTCH pathway alterations is still relatively new, several studies have assessed the frequencies of molecular events globally. These have demonstrated that 22/51 (43.1%) Chinese oral cavity HNSCC tumors harbored NOTCH1 alterations, with at least half predicted to activate function [109], while only 8/84 (9.5%) Japanese oral cavity HNSCCs had mutations that were all predicted to inactivate NOTCH1 activity [110]. In another cohort of patients from Singapore with tongue cancer (primarily Chinese but also Indian and Malay), NOTCH1 mutations occurred infrequently but alterations in other NOTCH pathway genes (i.e. AR, ARNT, EP300, CREBBP, JAK2, JAK3, NCOA1, NOTCH2, NOTCH3, and PARP1) were common (19/60, 32%) and correlated with disease survival [17]. While much larger cohort studies are needed, the preliminary published data indicates that activating NOTCH pathway alterations may be much more common in Chinese HNSCCs than in patients represented in the Indian ICGC and TCGA HNSCC projects.
Further studies examining the epidemiology of head and neck tumors with NOTCH pathway mutations will better characterize the role of these genetic aberrations in HNSCC. Rettig et al., based on an analysis of 79 tumor samples (primarily from Caucasian patients), reported that strong staining for the transcriptionally active NOTCH1 intracellular domain was more frequently observed in HPV-positive tumors. Mutations in NOTCH1 were more common in HPV-negative cases. In this study there was no difference between these NOTCH pathway alterations when comparing patients based on tobacco and alcohol use [111]. Similar analyses should be performed for other cohorts to confirm these findings and better understand the pathogenesis of the NOTCH wild-type and mutated head and neck cancers.
The Tumor Suppressor Protein, TP53
The p53 protein functions as a master regulator of the interplay between the cell cycle and apoptosis and is the most frequently deregulated tumor suppressor in HNSCC. In fact, the function and role of p53 in HNSCC have been reviewed extensively due to the high frequency of genetic or biochemical inactivation in the disease [112, 113]. In HPV-negative HNSCC, TP53 is commonly inactivated by mutation or deletion [114], while the HPV oncoproteins inactivate p53 by biochemical mechanisms in HPV-positive HNSCC. Thus, because TP53 is usually wild type in HPV-positive HNSCCs, we will restrict the review of genetic events for this gene to oral cavity and larynx HNSCCs, which are historically largely HPV-negative. At these sites, TP53 mutation often correlates with poorer survival [115] and has been associated with exposure to tobacco or betel quid [114]. Consequently, we may expect to find different rates of TP53 disruption in different epidemiologic subgroups if the gene is related to these risk factors, or may observe high rates across all populations if inactivation is generally required for squamous pathogenesis.
In the TCGA data set, TP53 mutation was observed in 129/160 (80.6%) HPV-negative tumors of the oral cavity [1, 77, 78]. In tongue cancer samples from a Singaporean cohort, TP53 was the most frequently mutated gene (as in TCGA), but was mutated in only 23/60 (38.3%) cases. Relatively low rates of TP53 mutation are consistent across multiple studies of oral cavity cancer in Asian patients [17, 116, 117]. Cohorts of Icelandic and American never-smokers also displayed lower oral cancer TP53 mutation rates [118, 119], while rates in Brazil and India were more consistent with those in the HPV-negative TCGA cohort [18, 120, 121]. Many of the patients in the ICGC study had HPV-positive, TP53 mutant tumors, which is inconsistent with previous studies showing no difference or lower mutation rates in HPV-positive HNSCC patients [122-124]. Other risk factors, such as tobacco use in addition to HPV infection, could contribute to this. Patients with multiple etiological factors are likely to have higher mutation rates and have also been shown to display decreased survival [46]. Further studies of HPV-positive HNSCC in this geographic (Singaporean) or anatomical (oral cavity) subset might better explain these differences.
Global TP53 mutation rates for oral cavity and larynx HNSCC are summarized in Tables 1 and 2, respectively. In the studies published thus far, TP53 mutation rates for laryngeal cancers are generally moderately higher than those in oral cavity cancer and also vary by geographic region. For example, in the TCGA cohort, 64/72 (88.9%) HPV-negative patients with laryngeal cancer display TP53 mutation [1, 77, 78]. Most other countries also have mutation rates of 50% or higher with the exception of China, South Africa, and Argentina [124-126]. Overall, however, these metadata are indicative of a relationship between inactivation and carcinogen exposure as opposed to specific pathogenic requirements.
Consistent with this observation, several groups have attempted to model the predictive value of individual TP53 mutations in different epidemiologic populations. For example, Ren et al. performed a meta-analysis to assess HNSCC risk and Arg72Pro TP53 mutation across various tumor sites in Asian and Caucasian cohorts. They found that this mutation was associated with increased risk of nasopharyngeal cancer, but not oral cancer, for homozygous, heterozygous and dominant model mutation comparisons in Caucasian cohorts but only homozygous mutations in Asian patients [127]. Given the frequency and complexity of TP53 aberration, further studies on the distinct role of this gene in specific epidemiological populations will be critical to developing an improved understanding of HNSCC pathogenesis.
FUTURE DIRECTIONS
Ultimately, additional sequencing of various epidemiologic sub-groups will need to be performed to understand the distribution of molecular events on a global scale. This work should also assess the correlations of disruptive genomic events with worldwide incidence, mortality, and particularly survival differences, which have not previously been taken into account. There is also a significant void of sequencing data in African populations in particular. Sequencing of these groups may identify both common and unique drivers for HNSCC between various cohorts. Studies of African populations are extremely important due to the high rate of HIV/AIDS patients in the region. We still have very limited knowledge of the pathogenesis or molecular distribution of HNSCCs in immunocompromised patients such as those who have HIV/AIDS or have undergone organ transplant. Additionally, analysis of cohorts worldwide will be necessary to determine the extent to which other environmental and genetic factors affect the incidence and severity of HNSCC both between and within epidemiologic sub-groups.
HPV-positive oropharyngeal squamous cell carcinoma now displays greater incidence than invasive cervical cancer [41]. While HPV vaccines have the potential to reduce the overall number of tumors caused by this virus, many populations around the world are incompletely vaccinated and it is unknown how many people encountered HPV before having the opportunity to be vaccinated. Thus, given the increasing rates of HPV in the US and abroad as well as wide variation between cohorts, there is a clear and urgent need for both epidemiologic data and sequencing analysis on HPV-positive tumors, especially those that are associated with additional risk factors. These studies have been performed partially, but include only analyses of HNSCC of the tongue or other regions of the oral cavity [17, 18] and not those cases involving more commonly HPV-positive oropharynx tumors. Fortunately, patients in the US with HPV-positive tumors are generally younger and have improved prognosis since HPV-positive tumors are more sensitive to chemoradiation [47]. Despite this fact, some HPV-positive tumors are highly aggressive and rapidly lethal, and there are no established biomarkers that can identify patients that would respond to more aggressive therapy. Future studies are needed to more fully elucidate the specific differences between HPV-positive patients from different geographic regions, cultural backgrounds, and genders using careful genetic analysis in the context of understanding the lethality of each tumor. These efforts are important as they may enable the development of biomarkers for the most aggressive forms of this epidemic subset of HNSCCs.
The heterogeneity of HNSCC has been clearly observed in previous studies and is dependent on tumor genetics and exposure to various risk factors including the use of tobacco, alcohol, and betel quid, HPV infection, and others. This variation is noted between not only between patients but also within individual tumors and between local, nodal, and distant tumor sites. Understanding the broad and underappreciated heterogeneity of HNSCC using comparative genetics will be valuable in establishing personalized medicine protocols first for clinical trials and then for individualized treatment plans. At this point, however, studies have not assessed the genetic heterogeneity found in individual tumors on a large scale. It is possible that different causal or associated factors driving HNSCC pathogenesis will lead to different levels of genetic heterogeneity within a tumor. As recurrence from targeted therapy can arise from individual cells with pre-existing resistant mutations, understanding the degree of heterogeneity in tumors from different epidemiologic subgroups may have a substantial impact on the choice of targeted therapy in each population.
Consequently, further sequencing analysis of patients will likely allow for more effective use of targeted therapies in countries where NGS analysis is readily available. Clinical trials are currently evaluating the response of HNSCC patients to single or dual targeted therapy treatments, such as EGFR (NCT02365662) or PI3K (NCT02145312, NCT02540928) monotherapies or both agents administered in combination (NCT0160231). However, these trials are rarely performed in subsets of patients with lesions that might make them more or less sensitive to particular treatment regimens. Specific genetic or clinicopathological characteristics may correlate with patient outcomes, but this effect will not be realized unless the genetic background of most (or all) of the individuals in the trial is known. One example of a trial that seeks to determine patient response in a subgroup of individuals with molecular enrichment is NCT02649530; this study is intended to evaluate WNT974 as a treatment for HNSCC patients with NOTCH1 deficient tumors.
Unfortunately, while precision-guided targeted antibodies and small molecule inhibitors display great promise in the future of cancer treatment, their use is currently limited by several disadvantages, including high cost and complex infusion regimens. Since genetic studies are perhaps most feasible in high income countries, understanding the contribution of epidemiological factors to HNSCC development and progression through various genetic pathways may enable treatments to be more effectively selected for patients. For example, understanding the high frequency genetic events in each region may restrict the number of biomarker tests needed to identify tumor drivers, and may enable clinicians to predict whether more or less aggressive therapy is needed based on those markers.
In the future, the hope is that access to healthcare resources and infrastructure will be enhanced globally so that patients have access to the best possible personalized therapy. In the meantime, we must think critically about the cost-benefit of biomarker-guided medicine in different epidemiologic subgroups of HNSCC in order to maximize the return on the high cost of NGS. Analyzing populations with unique epidemiologic and/or biomarker characteristics may be the first step to both enhancing our understanding globally and designing interventional protocols adapted to regional differences in health care resources and tumor genetics. We are in an exciting era of sequencing-guided personalized medicine in the US, and our challenge moving forward is to take the discoveries and lessons from these early personalized medicine trials, incorporate global sequencing information, and improve HNSCC therapy worldwide.
CONFLICT OF INTEREST
Dr. Brenner has previously collaborated with Novartis on the development of WNT974 for NOTCH-deficient HNSCC. The other authors have no conflicts of interest to disclose.
ACKNOWLEDGEMENTS
We acknowledge Elizabeth Leonard for reviewing this manuscript.
![Figure 2: Adjusted cervical HPV infection prevalence among women with normal cytology and oropharyngeal cancer incidence among men by geographic region [66, 69].](/article/110/asset/F2_thumb.png)
![Figure 3: Prevalence of key genetic aberrations in 24 black and 208 white HPV-negative patients (TCGA HNSCC cohort), 37 HPV-negative Indian patients (ICGC HNSCC cohort), 16 Indian HPV-negative patients (Krishnan cohort), and 60 patients from Singapore of unidentified HPV status (Vettore cohort) [1, 16-18].](/article/110/asset/F3_thumb.png)
- 1. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576-82. [PubMed] https://doi.org/10.1038/nature14129.
- 2. The Tip of the Iceberg: Clinical Implications of Genomic Sequencing Projects in Head and Neck Cancer. Cancers (Basel). 2015;7(4):2094-109. [PubMed] https://doi.org/10.3390/cancers7040879.
- 3. Getting personal: Head and neck cancer management in the era of genomic medicine. Head Neck. 2016;38 Suppl 1:E2250-8. doi: 10.1002/hed.24132. [PubMed] https://doi.org/10.1002/hed.24132.
- 4. Personalizing medicine in head and neck squamous cell carcinoma: The rationale for combination therapies. Med Res Arch. 2015;3. [PubMed] https://doi.org/10.18103/mra.v0i3.77.
- 5. NCI-MATCH launch highlights new trial design in precision-medicine era. J Natl Cancer Inst. 2015;107(7). [PubMed]
- 6. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324-34. [PubMed]
- 7. TAPUR Study Achieves Milestones: Forty-Nine Patients Consent to Participate, Bayer and Merck Sign on as Drug Providers, and Thirty-Seven Sites Are Participating Twelve Weeks Post-Launch. American Society of Clincal Oncology.. 2015;16(13):1324-34.
- 8. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70(13):5207-12. [PubMed] https://doi.org/10.1158/0008-5472.CAN-09-4074.
- 9. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22(5):806-21. [PubMed] https://doi.org/10.1038/cr.2012.30.
- 10. Targeted sequencing of an epidemiologically low risk patient defines Fibroblast Growth Factor Receptor family aberrations as a putative driver of head and neck squamous cell carcinoma. Head Neck. 2016; 38(Suppl 1): E1646–E1652.
- 11. Cancer statistics for African Americans, 2013. CA Cancer J Clin. 2013;63(3):151-66. [PubMed]
- 12. Unequal burden of head and neck cancer in the United States. Head Neck. 2008;30(3):358-71. [PubMed]
- 13. Disparities in oral and pharyngeal cancer incidence, mortality and survival among black and white Americans. J Am Dent Assoc. 2006;137(2):203-12. [PubMed]
- 14. Racial disparity in survival of patients with squamous cell carcinoma of the oral cavity and pharynx. Ethn Health. 2001;6(3-4):165-77. [PubMed]
- 15. Lifetime risks of cardiovascular disease. N Engl J Med. 2012;366(4):321-9. [PubMed] https://doi.org/10.1056/NEJMoa1012848.
- 16. Integrated analysis of oral tongue squamous cell carcinoma identifies key variants and pathways linked to risk habits, HP, clinical parameters and tumor recurrence. F1000Research. 2015;4:1215. [PubMed] https://doi.org/10.12688/f1000research.7302.1.
- 17. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome Med. 2015;7(1):98. [PubMed] https://doi.org/10.1186/s13073-015-0219-2.
- 18. Nat Commun. 2013;4:2873 [PubMed] https://doi.org/10.1038/ncomms3873.
- 19. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2008;83(4):489-501. [PubMed]
- 20. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18(2):541-50.
- 21. International trends in head and neck cancer incidence rates: differences by country, sex and anatomic site. Oral Oncol. 2014;50(5):387-403. [PubMed]
- 22. Systematic review and meta-analysis of association of smokeless tobacco and of betel quid without tobacco with incidence of oral cancer in South Asia and the Pacific. PLoS One. 2014;9(11):e113385. [PubMed] https://doi.org/10.1371/journal.pone.0113385.
- 23. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74-108. [PubMed]
- 24. Comparison of cancers of the oral cavity and pharynx worldwide: etiological clues. Oral Oncol. 2000;36(1):106-15. [PubMed]
- 25. Review: Head and neck squamous cell carcinoma in sub-Saharan Africa. Malawi Med J. 2015;27(3):79-87. [PubMed]
- 26. Current evidence on the burden of head and neck cancers in Nigeria. Head Neck Oncol. 2009;1:14. [PubMed] https://doi.org/10.1186/1758-3284-1-14.
- 27. Cancer incidence in Ghana, 2012: evidence from a population-based cancer registry. BMC Cancer. 2014;14:362. [PubMed] https://doi.org/10.1186/1471-2407-14-362.
- 28. Profile of cancer patients’ seen at Korle Bu teaching hospital in Ghana (a cancer registry review). BMC Res Notes. 2014;7:577. [PubMed] https://doi.org/10.1186/1756-0500-7-577.
- 29. Incidence of types of cancer among HIV-infected persons compared with the general population in the United States, 1992-2003. Ann Intern Med. 2008;148(10):728-36. [PubMed]
- 30. A review of the epidemiology of oral and pharyngeal carcinoma: update. Head Neck Oncol. 2012;4:1. [PubMed] https://doi.org/10.1186/1758-3284-4-1.
- 31. Ethnic differences in the occurrence of oropharyngeal cancer in Taiwan. Public Health. 2007;121(10):765-73. [PubMed]
- 32. K. Ethnic variation in CYP2E1, GSTM1, GSTT1 genes polymorphism analysis: Taiwan aborigines, Thai and Filipino. Mid Taiwan J Med. 2000;5:181-8.
- 33. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356(19):1944-56. [PubMed]
- 34. Clinical implications of human papillomavirus in head and neck cancers. J Clin Oncol. 2006;24(17):2606-11. [PubMed] https://doi.org/10.1200/JCO.2006.06.1291.
- 35. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100(6):407-20. [PubMed]
- 36. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):70920. [PubMed]
- 37. EGFR, p16, HPV Titer, BclxL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26(19):3128-37. [PubMed] https://doi.org/10.1200/JCO.2007.12.7662.
- 38. A multidisciplinary approach to squamous cell carcinomas of the head and neck: what is new? Curr Opin Oncol. 2006;18(3):253-7 [PubMed]
- 39. Tobacco use in human papillomavirus-positive advanced oropharynx cancer patients related to increased risk of distant metastases and tumor recurrence. Clin Cancer Res. 2010;16(4):122635. [PubMed] https://doi.org/10.1158/1078-0432.CCR-09-2350.
- 40. Chemoselection as a strategy for organ preservation in advanced oropharynx cancer: response and survival positively associated with HPV16 copy number. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(19):3138-46. [PubMed] https://doi.org/10.1200/JCO.2007.12.7597.
- 41. A comprehensive analysis of HPV integration loci in anogenital lesions combining transcript and genome-based amplification techniques. Oncogene. 2003;22(25):3977-84. [PubMed]
- 42. Detection of human papillomavirus type 16 integration in pre-neoplastic cervical lesions and confirmation by DIPS-PCR and sequencing. J Clin Virol. 2007;38(1):7-13. [PubMed]
- 43. Diagnosing cervical cancer and high-grade precursors by HPV16 transcription patterns. Cancer Res. 2010;70(1):249-56. [PubMed]
- 44. Transoral robotic surgery and adjuvant therapy for oropharyngeal carcinomas and the influence of p16 INK4a on treatment outcomes. Laryngoscope. 2013;123(3):635-40. [PubMed]
- 45. Transoral laser microsurgery as primary treatment for advanced-stage oropharyngeal cancer: a United States multicenter study. Head Neck. 2011;33(12):1683-94. [PubMed]
- 46. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24-35. [PubMed] https://doi.org/10.1056/NEJMoa0912217.
- 47. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261-9. [PubMed]
- 48. De-escalation treatment protocols for human papillomavirus-associated oropharyngeal squamous cell carcinoma: a systematic review and meta-analysis of current clinical trials. Eur J Cancer. 2014;50(15):2636-48. [PubMed]
- 49. Prognostic significance of p16 and its relationship with human papillomavirus in pharyngeal squamous cell carcinomas. JAMA otolaryngology-- head & neck surgery. 2014;140(7):647-53. [PubMed]
- 50. What is the role of human papillomavirus testing in head and neck cancer? Laryngoscope. 2015;125(12):2633-4 [PubMed]
- 51. Prevalence of Human Papillomavirus in Oropharyngeal Cancer: A Systematic Review. Cancer J. 2015;21(3):138-46. [PubMed] https://doi.org/10.1097/PPO.0000000000000115.
- 52. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer--systematic review and meta-analysis of trends by time and region. Head Neck. 2013;35(5):747-55. [PubMed]
- 53. Human papillomavirus positivity predicts favourable outcome for squamous carcinoma of the tonsil. Int J Cancer. 2003;106(4):553-8. [PubMed]
- 54. Prevalence, Clinicopathological Characteristics, and Outcome of Human Papillomavirus-Associated Oropharyngeal Cancer in Southern Chinese Patients. Cancer Epidemiol Biomarkers Prev. 2016; 25(1):165-73. [PubMed]
- 55. Prevalence of oral and oropharyngeal human papillomavirus in a sample of South African men: a pilot study. S Afr Med J. 2014;104(5):358-61. [PubMed]
- 56. Investigation of the presence of HPV related oropharyngeal and oral tongue squamous cell carcinoma in Mozambique. Cancer Epidemiol. 2015;39(6):1000-5. [PubMed] https://doi.org/10.1016/j.canep.2015.10.015.
- 57. The role of human papillomavirus in head and neck cancer in Senegal. Infect Agent Cancer. 2013;8(1):14. [PubMed] https://doi.org/10.1186/1750-9378-8-14.
- 58. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med. 2001;344(15):1125-31. [PubMed]
- 59. Detection of human papillomavirus (HPV) in clinical samples: evolving methods and strategies for the accurate determination of HPV status of head and neck carcinomas. Oral Oncol. 2014;50(9):771-9. [PubMed] https://doi.org/10.1016/j.oraloncology.2014.05.004.
- 60. p16 Immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2012;6 Suppl 1:S75-82. [PubMed] https://doi.org/10.1007/s12105-012-0369-0.
- 61. Highrisk human papillomavirus detection in oropharyngeal, nasopharyngeal, and oral cavity cancers: comparison of multiple methods. JAMA otolaryngology-- head & neck surgery. 2013;139(12):1320-7. [PubMed] https://doi.org/10.1001/jamaoto.2013.5460.
- 62. Prevalence of oral HPV infection in the United States, 2009-2010. JAMA. 2012;307(7):693-703. [PubMed] https://doi.org/10.1001/jama.2012.101.
- 63. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol Oncol. 2009;113(1):120-7. [PubMed]
- 64. Age-specific prevalence of infection with human papillomavirus in females: a global review. J Adolesc Health. 2008;43(4 Suppl):S5-25, S.e1-41. [PubMed]
- 65. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30 Suppl 5:F12-23. [PubMed]
- 66. Cervical human papillomavirus prevalence in 5 continents: meta-analysis of 1 million women with normal cytological findings. J Infect Dis. 2010;202(12):1789-99. [PubMed]
- 67. Age-specific prevalence of human papillomavirus infection in males: a global review. J Adolesc Health. 2011;48(6):540-52. [PubMed]
- 68. Male circumcision, penile human papillomavirus infection, and cervical cancer in female partners. N Engl J Med. 2002;346(15):1105-12. [PubMed]
- 69. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550-9. [PubMed] https://doi.org/10.1200/JCO.2013.50.3870.
- 70. Sexual behaviours and the risk of head and neck cancers: a pooled analysis in the International Head and Neck Cancer Epidemiology (INHANCE) consortium. Int J Epidemiol. 2010;39(1):166-81. [PubMed] https://doi.org/10.1093/ije/dyp350.
- 71. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J Clin Oncol. 2015;33(29):3235-42. [PubMed] https://doi.org/10.1200/JCO.2015.61.6995.
- 72. Differences in oral sexual behaviors by gender, age, and race explain observed differences in prevalence of oral human papillomavirus infection. PLoS One. 2014;9(1):e86023. [PubMed] https://doi.org/10.1371/journal.pone.0086023.
- 73. NHANES 2009-2012 Findings: Association of Sexual Behaviors with Higher Prevalence of Oral Oncogenic Human Papillomavirus Infections in U.S. Men. Cancer Res. 2015;75(12):2468-77. [PubMed] https://doi.org/10.1158/0008-5472.CAN-14-2843.
- 74. EUROGIN 2014 roadmap: differences in human papillomavirus infection natural history, transmission and human papillomavirus-related cancer incidence by gender and anatomic site of infection. Int J Cancer. 2015;136(12):2752-60. [PubMed] https://doi.org/10.1002/ijc.29082.
- 75. Racial survival disparity in head and neck cancer results from low prevalence of human papillomavirus infection in black oropharyngeal cancer patients. Cancer Prev Res (Phila). 2009;2(9):776-81. [PubMed] https://doi.org/10.1158/1940-6207.CAPR-09-0149.
- 76. Racial disparities in Human Papillomavirus (HPV) associated head and neck cancer. Am J Otolaryngol. 2014;35(2):147-53. [PubMed] https://doi.org/10.1016/j.amjoto.2013.09.004.
- 77. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-4. [PubMed] https://doi.org/10.1158/2159-8290.CD-12-0095.
- 78. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [PubMed] https://doi.org/10.1126/scisignal.2004088.
- 79. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157-60. [PubMed] https://doi.org/10.1126/science.1208130.
- 80. Over-expression of the EGF receptor is a hallmark of squamous cell carcinomas. J Pathol. 1986;149(1):9-14. [PubMed]
- 81. Platinum based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116-27.
- 82. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354(6):567-78. [PubMed]
- 83. Role of EGFR as a prognostic factor for survival in head and neck cancer: a meta-analysis. Tumour Biol. 2014;35(3):2285-95. [PubMed]
- 84. Expression of EGFR and p53 in head and neck tumors among Sudanese patients. Asian Pac J Cancer Prev. 2013;14(11):6415-8. [PubMed]
- 85. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11(8):2879-82. [PubMed]
- 86. Low incidence of mutations in EGFR kinase domain in Caucasian patients with head and neck squamous cell carcinoma. Eur J Cancer. 2006;42(1):109-11. [PubMed]
- 87. Identification of the rare EGFR mutation p.G796S as somatic and germline mutation in white patients with squamous cell carcinoma of the head and neck. Head Neck. 2008;30(8):1040-4. [PubMed]
- 88. Absence of activating mutations in the EGFR kinase domain in Spanish head and neck cancer patients. Tumour Biol. 2007;28(5):273-9. [PubMed]
- 89. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12(17):5064-73. [PubMed]
- 90. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761-9. [PubMed] https://doi.org/10.1158/2159-8290.CD-13-0103.
- 91. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9(6):667-76. [PubMed]
- 92. Targeting the PI3K Pathway in Head and Neck Squamous Cell Carcinoma. Am Soc Clin Oncol Educ Book. 2015:123-8. [PubMed]
- 93. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(3):632-41. [PubMed] https://doi.org/10.1158/1078-0432.CCR-13-3310.
- 94. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58(3):509-11. [PubMed]
- 95. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(5):1441-6. [PubMed] https://doi.org/10.1158/1078-0432.CCR-05-2173.
- 96. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006;97(12):1351-8. [PubMed]
- 97. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol. 2008;32(1):101-11. [PubMed]
- 98. Analysis of PIK3CA and Akt/protein kinase B in head and neck squamous cell carcinoma. Oncol Rep. 2007;18(1):2539. [PubMed]
- 99. Absence of hotspot mutations in exons 9 and 20 of the PIK3CA gene in human oral squamous cell carcinoma in the Greek population. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;109(5):e538. [PubMed]
- 100. Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous cell carcinoma. Oral Oncol. 2011;47(10):946-50. [PubMed]
- 101. Copy number amplification of the PIK3CA gene is associated with poor prognosis in non-lymph node metastatic head and neck squamous cell carcinoma. BMC Cancer. 2012;12:416. [PubMed] https://doi.org/10.1186/1471-2407-12-416.
- 102. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001;61(10):4122-9. [PubMed]
- 103. Genetic deregulation of the PIK3CA oncogene in oral cancer. Cancer Lett. 2013;338(2):193-203. [PubMed]
- 104. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154-7. [PubMed] https://doi.org/10.1126/science.1206923.
- 105. Activation of the NOTCH pathway in head and neck cancer. Cancer research. 2014;74(4):1091-104. [PubMed] https://doi.org/10.1158/0008-5472.CAN-13-1259.
- 106. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649-61. [PubMed]
- 107. The opposing roles of NOTCH signalling in head and neck cancer: a mini review. Oral Dis. 2015;21(7):850-7. [PubMed]
- 108. In vivo Wnt pathway inhibition of human squamous cell carcinoma growth and metastasis in the chick chorioallantoic model. J Otolaryngol Head Neck Surg. 2016;45:26. [PubMed] https://doi.org/10.1186/s40463-016-0140-8.
- 109. Common and complex Notch1 mutations in Chinese oral squamous cell carcinoma. Clin Cancer Res. 2014;20(3):70110. [PubMed] https://doi.org/10.1158/1078-0432.CCR-13-1050.
- 110. Frequent mutations in NOTCH1 ligand-binding regions in Japanese oral squamous cell carcinoma. Biochem Biophys Res Commun. 2014;452(4):980-5. [PubMed]
- 111. Cleaved NOTCH1 Expression Pattern in Head and Neck Squamous Cell Carcinoma Is Associated with NOTCH1 Mutation, HPV Status, and High-Risk Features. Cancer Prev Res (Phila). 2015;8(4):287-95. [PubMed] https://doi.org/10.1158/1940-6207.CAPR-14-0366.
- 112. TP53 and head and neck neoplasms. Hum Mutat. 2003;21(3):252-7. [PubMed]
- 113. The p53 network in head and neck cancer. Oral Oncol. 2003;39(3):222-31. [PubMed]
- 114. A pilot study evaluating genetic alterations that drive tobacco- and betel quid-associated oral cancer in Northeast India. Tumour Biol. 2014;35(9):9317-30. [PubMed]
- 115. Combined mutational analysis of RAS, BRAF, PIK3CA, and TP53 genes in Taiwanese patients with oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;118(1):110-6. [PubMed]
- 116. Tongue carcinoma infrequently harbor common actionable genetic alterations. BMC Cancer. 2014;14:679. [PubMed] https://doi.org/10.1186/1471-2407-14-679.
- 117. Common oncogenic mutations are infrequent in oral squamous cell carcinoma of Asian origin. PLoS One. 2013;8(11):e80229. [PubMed] https://doi.org/10.1371/journal.pone.0080229.
- 118. Oral lichen planus has a high rate of TP53 mutations. A study of oral mucosa in icelanD. Eur J Oral Sci. 2002;110(3):192-8. [PubMed]
- 119. TP53 and CDKN2a mutations in never-smoker oral tongue squamous cell carcinoma. Laryngoscope. 2014;124(7):E267-73. [PubMed]
- 120. TP53 mutations in upper aerodigestive squamous cell carcinomas from a group of Brazilian patients. Am J Surg. 1995;170(5):492-4. [PubMed]
- 121. Characterization of p53 gene mutations in a Brazilian population with oral squamous cell carcinomas. Int J Oncol. 2004;24(2):295-303. [PubMed]
- 122. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25(17):2558-64. [PubMed]
- 123. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96(13):998-1006. [PubMed]
- 124. Associations of TP53 mutations, codon 72 polymorphism and human papillomavirus in head and neck squamous cell carcinoma patients. Oncol Rep. 2013;30(6):2811-9. [PubMed]
- 125. The spectrum of mutations in TP53 in laryngeal cancer patients from a high-incidence population shows similarities to many of the known mutational hotspots. Cancer Genet Cytogenet. 2003;145(2):126-32. [PubMed]
- 126. TP53 and EGFR mutations in combination with lifestyle risk factors in tumours of the upper aerodigestive tract from South America. Carcinogenesis. 2010;31(6):1054-9. [PubMed]
- 127. Meta-analysis of associations between the TP53 Arg72Pro polymorphism with risk of head and neck carcinomas based on case-control studies. Genet Mol Res. 2014;13(1):103-14. [PubMed]
- 128. Survivin and laryngeal carcinoma prognosis: nuclear localization and expression of splice variants. Histopathology. 2012;61(2):247-56. [PubMed]
- 129. TP53 mutations and S-phase fraction but not DNAploidy are independent prognostic indicators in laryngeal squamous cell carcinoma. J Cell Physiol. 2006;206(1):1818. [PubMed]
- 130. Molecular profiles as predictive marker for the effect of overall treatment time of radiotherapy in supraglottic larynx squamous cell carcinomas. Radiother Oncol. 2004;72(3):275-82. [PubMed]
- 131. Trends in the incidence of cancer in the black population of Harare, Zimbabwe 1991-2010. Int J Cancer. 2013;133(3):721-9. [PubMed]
- 132. Trends of lip, oral cavity and oropharyngeal cancers in Australia 1982-2008: overall good news but with rising rates in the oropharynx. BMC Cancer. 2013;13:333. [PubMed] https://doi.org/10.1186/1471-2407-13-333.
- 133. Trends in the incidence of Oral and Pharyngeal Cancer (ICD00-14) in Guilan, North of Iran. J Oral Pathol Med. 2016; 45(4):275-80. [PubMed]
- 134. Exploiting highthroughput cell line drug screening studies to identify candidate therapeutic agents in head and neck cancer. BMC Pharmacol Toxicol. 2014;15:66. [PubMed] https://doi.org/10.1186/2050-6511-15-66.
- 135. Genetic alterations of the PIK3CA oncogene in human oral squamous cell carcinoma in an Indian population. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015;120(5):628-35. [PubMed]
- 136. Molecular characterization of oral squamous cell carcinoma using targeted next-generation sequencing. Oral Dis. 2015;21(7):872-8. [PubMed]
- 137. Ultra-deep targeted sequencing of advanced oral squamous cell carcinoma identifies a mutation-based prognostic gene signature. Oncotarget. 2015;6(20):18066-80. [PubMed] https://doi.org/10.18632/oncotarget.3768.
- 138. Nature. 2015;517(7536):576-82
- 139. Receptor tyrosine kinase profiles and human papillomavirus status in oropharyngeal squamous cell carcinoma. J Oral Pathol Med. 2015;44(9):734-45. [PubMed]
- 140. Genetic alterations in oral squamous cell carcinoma progression detected by combining array-based comparative genomic hybridization and multiplex ligation-dependent probe amplification. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011;111(5):594-607. [PubMed]
- 141. The frequent co-expression of the oncogenes PIK3CA and PAK1 in oral carcinomas. Oral Oncol. 2011;47(3):2116. [PubMed]
Last Modified: 2016-07-28 20:34:08 EDT