Genes & Cancer
TET1-mediated DNA hypomethylation regulates the expression of MUC4 in lung cancer
Seiya Yokoyama1,2,3, Michiyo Higashi1,2, Hideaki Tsutsumida1, Jouji Wakimoto4, Tomofumi Hamada5, Edwin Wiest3, Kei Matsuo1, Ikumi Kitazono1, Yuko Goto1, Xin Guo1, Taiji Hamada1, Sohsuke Yamada1, Tsubasa Hiraki1, Suguru Yonezawa1, Surinder K. Batra6, Michael A. Hollingsworth3, Akihide Tanimoto1
1 Department of Pathology, Research Field in Medicine and Health Sciences, Medical and Dental Sciences Area, Research and Education Assembly, Kagoshima University, Sakuragoaka, Japan
2 Center for the Research of Advanced Diagnosis and Therapy of Cancer, Graduate School of Medical and Dental Sciences, Kagoshima University, Japan
3 Eppley Institute for Research in Cancer, Fred and Pamela Buffet Cancer Center, University of Nebraska Medical Center, NE, USA
4 Department of Respiratory Medicine, Minami-kyushu National Hospital, Aira, Japan
5 Department of Oral Surgery, Kagoshima University Medical and Dental Hospital, Sakuragoaka, Japan
6 Department of Biochemistry and Molecular Biology, Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, NE, USA
Correspondence to: Michiyo Higashi, email: [email protected]
Keywords: Lung cancer, pathology, DNA methylation, risk factor, MUC4
Received: April 25, 2017
Accepted: June 14, 2017
Published: June 19, 2017
Copyright: Yokoyama et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Lung cancer remains a disease of high mortality, despite advanced diagnostic techniques. Mucins (MUC) play crucial roles in carcinogenesis and tumor invasion in lung neoplasms. Our immunohistochemistry (IHC) studies have shown that high MUC4 expression correlates with a poor outcome. We have also shown that the expression of several mucin genes in cancer cell lines is regulated by DNA methylation. We evaluated the expression level of MUC4, mRNA and several DNA hypomethylation factors in lung tissue samples from 33 patients with various lung lesions. The results indicated that the DNA methylation status of MUC4 matched the expression level of mRNA. In addition, the TET1 (Ten-Eleven Translocation) mRNA showed a significant correlation with the status of DNA methylation of MUC4. Furthermore, the treatment of a lung cancer cell line with TET1 siRNA caused a reduction in MUC4 mRNA expression. Thus, we suggest that TET1 mediated DNA hypomethylation plays a key role in the expression of MUC4. This is the first report that TET1 mediated DNA hypomethylation regulates the expression of MUC4 in lung cancer. The analysis of these epigenetic changes may be useful for diagnosing carcinogenic risk.
INTRODUCTION
Lung cancer is the leading cause of cancer-related death in most industrialized countries [1, 2], and adenocarcinomas account for approximately 45% of lung cancer [3]. Poor prognosis for patients with lung cancer has persisted, despite efforts in primary prevention, screening and therapy [1]. Within current screening techniques for patients that lack symptoms, a diagnostic method to differentiate small lung adenocarcinomas from benign lesions is needed.
Mucins (MUC) play crucial roles in carcinogenesis and tumor invasion in lung neoplasms. MUC4, a large membrane-bound glycoprotein that is translated as a single polypeptide, undergoes intracellular autocatalytic proteolytic cleavage into two subunits that form a stable non-covalent heterodimer that is transported to the cell surface. MUC4 plays an important role in cell proliferation and differentiation of epithelial cells by inducing specific phosphorylation of ErbB2 and enhancing the expression of the cyclin dependent kinase inhibitor p27, which inhibits cell cycle progression [4-11]. Our immunohistochemistry (IHC) studies have revealed that a high MUC1/SP-A ratio is strongly associated with a poor outcome in patients with small-size lung adenocarcinoma and that high MUC4 expression in lung adenocarcinoma patients associates with poor outcome [12-14]. We have also found that the methylation status, mRNA expression, and protein expression of mucins in cancer cell lines are correlated [15-17]. We have shown that mucin gene expression is regulated by DNA methylation status in pancreatic tissue [18, 19]. In addition, we reported that hypomethylation of the MUC4 promoter correlates with a decreased overall survival in pancreatic ductal adenocarcinoma [20].
Bisulfite treatment is a current standard for DNA methylation analysis. However, one pitfall with the bisulfite treatment is that 5-hydroxy methyl cytosine (5hmC) is detected as 5-methyl cytosine (5mC). 5hmC is the primary product of 5mC oxidation, a process that plays an essential role in normal embryonic development and the maintenance of pluripotency and stem cell reprogramming [21-24]. Recently, it was reported that not only DNA methylated by the Dnmt (DNA methyltransferase) family but also DNA modified by TET (Ten-Eleven Translocation) family, AICDA (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) family and/or GCM1 (glial cells missing 1) convert 5mC to 5hmC and higher oxidation products in mammalian genomes (i.e. active DNA hypomethylation) [25-31].
In this study, we sought to further characterize the epigenetic changes of the MUC4 promoter region in lung adenocarcinomas through analysis of DNA samples with the MSE method (with bisulfite treatment and/or TET assisted bisulfite treatment). As no recent study has evaluated the extent of 5hmC modification of the MUC4 gene and correlated this to expression levels of MUC4 mRNA in lung tumors, we analyzed MUC4 5mC status and/or 5hmC status in lung tissue to study the relationship between MUC4 promoter modification and expression.
RESULTS
Correlation between DNA methylation status and mRNA expression.
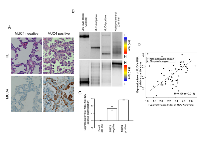
In total, 66 lung tissue samples were collected from 33 lung cancer patients (Table S1). We examined the relationship between MUC4 mRNA expression, DNA methylation of the promoter, and IHC staining for MUC4 protein in paired lung tissues. Representative cases of mRNA expression (RT-PCR) paired with IHC analysis and 5mC score and 5hmC score are shown in Figure 1. We found that IHC positive samples were mRNA positive and that IHC negative samples were mRNA negative (Figure 1A and 1C). We observed similar methylation patterns of 5mC in both MUC4 positive and MUC4 negative lung tissues; however, 5hmC status was correlated with expression of MUC4 protein (Figure 1B). We analyzed the relationship between the 5mC or the 5hmC score of the MUC4 promoter and the expression level of MUC4 mRNA with Pearson’s correlation coefficient (R=-0.323, p=0.011 and R=0.105, p=0.426, Table 1). A significant degree of correlation was observed between the hypomethylation index (calculated by the following formula: hypomethylation index = 2.94+(1.32(5hmC score)-0.98(5mC score))/1000) and mRNA expression of MUC4 (R=0.326, p=0.001, Figure 1D).
Differences in methylation status between neoplastic and non-neoplastic areas.
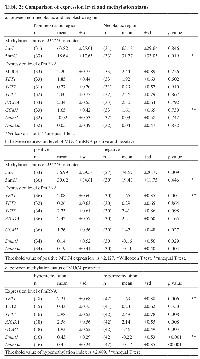
Thirty-three neoplastic samples and 33 paired non-neoplastic samples were analyzed. No significant difference was observed for expression of MUC4 mRNA in neoplastic tissues versus non-neoplastic tissues. However, there was a statistically significant difference in MUC4 mRNA expression in samples positive or negative for MUC4 (as determined by IHC analysis) (p=0.013, Supplemental Figure 1A). A threshold value of MUC4 mRNA expression that could distinguish between positive and negative MUC4 IHC staining was determined to be 2.127 by ROC analysis (Supplemental Figure 1B). A dot-blot analysis was used to examine differences in 5hmC modification of the MUC4 gene between neoplastic and non-neoplastic regions obtained from lung tissues (Figure 2A). Non-neoplastic areas showed a significantly higher level of 5hmC than neoplastic areas (p=0.020, Figure 2B). On the other hand, 5hmC modification of the MUC4 promoter region in non-neoplastic regions was lower than in neoplastic regions (P=0.019, Figure 2C). There was no significant difference in 5hmC modification of the MUC4 promoter region between neoplastic and non-neoplastic regions. However, within the MUC4 mRNA negative group, higher levels of 5mC modification were observed compared to that of the MUC4 mRNA positive group (p=0.009, Figure 2D). These data are summarized in Table 2. These results suggest that, including 5hmC, the neoplastic area has an increased hypomethylation status in the MUC4 promoter region. However, overall 5hmC modification within the neoplastic areas was lower than in the non-neoplastic areas.
5mC/5hmC score and expression of DNA methylation-related enzymes in lung tissue.
The mRNA expression levels of DNA methylation-related enzymes (DNMT1 and DNMT3a) and DNA demethylation-related enzymes (TET1, TET2, TET3, AICDA and GCM1) in neoplastic and non-neoplastic samples are summarized in Table 2. There were no differences in expression of these between neoplastic and non-neoplastic regions. However, a comparison between the MUC4 mRNA positive group and negative group revealed significant differences in expression levels of TET1, GCM1, Dnmt1 and Dnmt3a (p=0.001, p=0.027, p=0.029 and p=0.004, respectively). The expression level of TET1 showed a significant correlation with the expression level of MUC4 (R=0.543, p<0.001, Table 1). To examine whether the MUC4 promoter hypomethylation is influenced by the expression of DNA methylation-related enzymes, we analyzed the expression level of these enzymes in the hypomethylated and hypermethylated groups. The threshold value of methylation index to distinguish between hypomethylation and hypermethylation of the MUC4 promoter was 2.489 as determined by ROC analysis (Supplemental Figure 1C and 1D). The hypomethylated group showed higher expression levels of TET1, TET3, GCM1, AICDA, Dnmt1 and Dnmt3a than the hypermethylated group (Table 2). The expression level of TET1, TET2, AICDA, Dnmt1 or Dnmt3a correlated with the hypomethylation index (R=0.392, R=0.388, R=0.385, R=0.551 and R=0.636, respectively, Table 1). In order to find statistically significant differences between enzymes related to DNA methylation, we performed a multiple regression analysis. We determined the best regression formula with the least variables (five DNA demethylation-related enzymes) with the lowest AIC values for the hypomethylation status of MUC4 as follows: Fm (Enzyme expression index for MUC4) = 1.8 + 0.23(TET1) + 0.17(TET2). This predictive model showed a significantly high correlation with the hypomethylation index of MUC4 (R2 = 0.562, p<0.001, Supplemental Figure 2).
Correlation between expression level of DNA methylation-related enzymes and hypomethylation status of MUC4 and clinicopathological features.
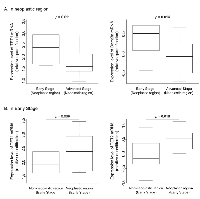
Expression levels of DNA methyltransferases (DNMTs) as DNA methylation factors (DNMT1 and DNMT3a), DNA demethylation factors (TET1, TET2, TET3, AICDA and GCM1) and MUC4 were evaluated in tumors representative of early stage (Tumor size < 10mm), later stages, lymphatic permeation negative and positive samples, and vascular permeation negative and positive samples (summarized in Table S3). Analysis of vascular permeation negative and positive samples revealed no significant differences in DNA methylation-related enzymes, MUC4 expression levels, or MUC4 methylation status. However, analysis of samples of the neoplastic region without lymphatic permeation showed higher expression of TET1, Dnmt1, and Dnmt3a than samples of the neoplastic region with lymphatic permeation (p=0.020, p=0.032 and p=0.005 respectively). In the case of samples with lymphatic permeation, the neoplastic region showed a higher 5hmC score in the MUC4 promoter than the paired non-neoplastic region (p=0.004). Early stage lung cancers showed higher expression of TET1 and Dnmt3a than other advanced stages (p=0.011 and p=0.014 respectively, Figure 3A). In early stage samples, the neoplastic region showed higher TET1 and TET2 expression than the paired non-neoplastic region (p=0.009 and p=0.016 respectively, Figure 3B).
Effect of TET1 knockdown on MUC4 expression in cancer cell lines.
To further explore a causal relationship between TET1 expression and activity and MUC4 expression, lung cancer cell lines (A427 and NCI-H292) were employed. When endogenous TET1 in A427 cells (MUC4 positive) was knocked down by siRNA (Figure 4A), the MUC4 expression level was strongly reduced (p=0.001). In contrast, siRNA knockdown of TET1 in the MUC4 negative NCI-H292 cell line was ineffective in changing MUC4 expression (Figure 4B). Also, knockdown of TET1 caused no change in MUC1 expression (Figure 4C). These data suggest that TET1 plays a key role in regulating the expression of MUC4 mRNA.
DISCUSSION
In the present study, we analyzed the correlation between MUC4 expression and DNA methylation and 5mC and/or 5hmC scores in the promoter region of MUC4 in lung adenocarcinomas. It has been shown previously that expression of mucin genes such as MUC1, MUC2, MUC3, MUC4, and MUC5AC are regulated by DNA methylation (5mC) of these promoter regions [15-17]. Our results are the first to demonstrate that TET1-mediated DNA hypomethylation regulates the expression of MUC4 in lung adenocarcinomas.
In our comparison of lung neoplastic and non-neoplastic samples, we found a significant difference in 5hmC scores in the MUC4 promoter region. However, the level of 5mC in the promoter of MUC4 showed no difference when the neoplastic non-neoplastic regions were compared, while the 5hmC score of the MUC4 promoter was increased in the neoplastic region when compared with the non-neoplastic region. In contrast, in whole DNA, the non-neoplastic region of the lung showed higher 5hmC scores when compared with the neoplastic region. Some recent studies have also shown that 5hmC is substantially decreased in human prostate, breast, colon, lung, liver, and pancreatic cancers, as well as glioma and melanoma [24, 32-36]. Therefore, our results show that alteration of the DNA hypomethylation process, such as that of the MUC4 promoter region, can be a gene-specific process that persists in spite of overall trends in hypomethylation. The expression level of MUC4 was not found to be different when comparing the neoplastic region to the non-neoplastic region. This may be because MUC4 is expressed only in neoplasms with poor outcome, as most lung tissue does not express MUC4. Thus, these results suggest that the DNA hypomethylation process, conversion of 5mC to 5hmC in the MUC4 promoter, precedes increases in expression of MUC4 in the lung neoplastic region.
Our evaluation of the relationship between MUC4 expression level and the degree of hypomethylation of MUC4 revealed that the group with high expression of MUC4 mRNA showed a higher 5mC score of MUC4 than the group with low expression of MUC4 mRNA. The hypomethylation score (calculated by comparison of 5mC and 5hmC scores) showed a significant correlation with the expression of MUC4 mRNA. These results complement the results in lung cancer cell lines that we found in our previous study [16] and suggest that MUC4 expression is regulated by epigenetic DNA modification (e.g., 5mC and/or 5hmC) in lung adenocarcinomas as well as in non-neoplastic lung tissue.
Concerning the relationship between MUC4 promoter hypomethylation and expression of several epigenetic alteration factors such as the TET family, AICDA/Apobec family, GCM1 and Dnmt family, we found significant differences between the MUC4 hypomethylated group and the MUC4 hypermethylated group. The expression levels of the active hypomethylation factors TET1, TET3, GCM1 and AICDA in the MUC4 hypomethylation group were significantly higher than in the MUC4 hypermethylation group. Similarly, the MUC4 hypomethylation group showed a significantly higher expression level of DNA methylation factors, Dnmt1 and Dnmt3a, than the MUC4 hypermethylation group. Multiple correlation analysis showed that the expression levels of TET1 and TET2 significantly correlated with the hypomethylation index of the MUC4 promoter. These results suggest that expression of the MUC4 gene is increased when 5mC levels and/or 5hmC modifications at the MUC4 promoter region are altered, and this alteration may be caused by activation of these DNA methylation-related enzymes.
A comparison of DNA methylation-related enzymes, MUC4 methylation status and clinicopathological information, revealed significantly higher expression of TET1, Dnmt1 and Dnmt3a in the neoplastic region with lymphatic permeation than in the neoplastic region without it. Interestingly, in our samples TET1 was downmodulated according to the tumor size. Moreover, TET1 expression was significantly higher in the early stage (tumor size < 10mm) neoplastic region than in the paired non-neoplastic region. This result suggests that TET1 expression is the initial step in reprogramming DNA methylation in lung cancer. In addition, we found a significant correlation between TET1 expression and MUC4 mRNA expression. We showed a significant reduction of MUC4 mRNA by TET1 mRNA down-regulation in a lung cancer cell line. We suggest that increased expression of TET1 may cause hypomethylation and/or the conversion of 5mC to 5hmC or higher oxidation products in the MUC4 promoter region. These results suggest that demethylation of the MUC4 promoter by TET1 may be involved in the early stage and/or in the production of precursor cancer cells of lung cancer.
In summary, our data demonstrate that MUC4 expression is increased by DNA hypomethylation when both 5mC and 5hmC are considered. Furthermore, MUC4 hypomethylation status is statistically associated with active methylation and/or hypomethylation factors. Moreover, in the early stage, TET1 plays a key role in MUC4 hypomethylation. Thus, detection of the hypomethylation index of MUC4 and these DNA methylation-related factors has potential clinical value as an indicator of overall survival and should be evaluated further for clinical utility. Since MUC4 is a key mucin in pathological diagnosis of lung neoplasms [12, 14], our goal is to apply DNA methylation analysis of this gene using bronchoalveolar lavage fluid and/or sputum for early diagnosis of lung neoplasms.
MATERIALS AND METHODS
Cell lines
Human lung carcinoma cell lines A427 and NCI-H292 were obtained from the American Type Culture Collection. A427 was cultured in Eagle’s minimum essential medium (Sigma, St. Louis, MO, USA), and NCI-H292 was cultured in RPMI 1640 medium (Sigma, St. Louis, MO, USA). The media was supplemented with 10% fetal bovine serum (Invitrogen, Minatoku, Tokyo, Japan) and 100 U/mL of penicillin and 100 μg/mL streptomycin (Sigma).
Clinical samples Lung tissue samples
We aimed to examine the relationship between the extent of DNA methylation of mucin genes and expression of mRNA in paired lung tissues. We obtained tissue blocks (about 2×2×2 mm) with neoplastic and non-neoplastic areas from surgically resected fresh specimens of 30 adenocarcinomas, 2 squamous cell carcinomas, and 1 adenosquamous carcinoma and paired non-neoplastic samples. Table S1 summarizes the clinicopathological characteristics of the samples analyzed herein.
Ethics statement
The study was conducted in accordance with the guiding principles of the Declaration of Helsinki. Collection of samples was approved by the ethical committees of the hospital and informed written consent was obtained from each patient. All studies using human materials in this article were approved by the Ethical Committee of Kagoshima University Hospital (revised 20-82, revised 22-127, and revised 26-145).
Extraction and Quantification of mRNA
Total RNA was extracted from cell lines and human lung tissues using an RNeasy Mini kit (QIAGEN, Tokyo, Japan). Total RNA (1 μg) was reverse transcribed with a high capacity RNA-to-cDNA Kit (Applied Biosystems, CA, USA). Real-time reverse transcription–PCR was performed on a Roche LightCycler® 96 System using FastStart Essential DNA Green Master (Roche, Tokyo, Japan). Gene expression was normalized to the β-actin mRNA level in each sample. The data of the A427 cell line were used for basal control. Primer sets are shown in Table S2.
Dot blot analysis
DNA was denatured in 0.4 M NaOH, 10 mM EDTA at 95°C for 10 minutes, and then neutralized by adding an equal volume of cold 2 M ammonium acetate (pH 7.0). Next, 2-fold dilutions of denatured DNA samples were spotted on a Hybond N+ nylon membrane. The DNA was fixed by UV cross-linking, washed with 2x SSC buffer and air-dried. The membrane was then blocked with 5% non-fat milk and incubated with polyclona 5hmC antibody (1:1000) (active motif). Binding of an HRP-conjugated secondary antibody (1:12,000) was visualized by enhanced chemiluminescence. The blot intensity was measured by Image J software (National Institutes of Health http://rsb.info.nih.gov/ij/). The dot blot intensity in each sample was normalized to the amount of DNA applied to the membrane.
Extraction of DNA and Bisulfite Modification
DNA from cell lines and lung tissue was extracted using a DNeasy Tissue System (QIAGEN). Bisulfite modification of the genomic DNA was carried out using an Epitect Bisulfite Kit (QIAGEN). Purification of PCR products was carried out using a Wizard SV Gel and PCR Clean-Up System (Promega).
TET1 assisted bisulfite (TAB) treatment
For measuring 5hmC in the MUC4 promoter, collected total DNA was treated by TAB treatment similar to that used by Yu et al. [36]. In this method, to protect 5hmC, the DNA sample was treated with β-glucosyltransferase. Subsequently, recombinant TET1 was used to convert 5mC to 5-formylcytosine (5-fC) and/or 5-carboxylcytosine (5-caC). After the bisulfite treatment and PCR amplification, both cytosine (C) and higher oxidation products of 5mC (i.e. 5-fC and 5-caC) are converted to thymine (T), whereas protected 5hmC remains C.
MSE Analysis
MSE analysis was performed using previously described methods [19]. Briefly, the target DNA fragments were amplified by nested PCR using bisulfite treated DNA using the primer sets shown in Table S2. In the electrophoresis step, the amplicon was applied to the D-Code system (BioRad Laboratories, Hercules, CA, USA) using a polyacrylamide gel with a linear denaturant gradient at 60°C and 70 V for 14 h. Band intensity was quantified by Image J software. The 5mC score and 5hmC score were calculated as the proportion of highest band intensity to total band intensity of the sample. Subsequently, these scores in each sample were normalized using data from a hypomethylated and hypermethylated control. Cell lines that are hyper- and hypomethylated (Caco-2 and LS174T) were used as controls for determination of 5mC scores. An oligonucleotide sequence (all CpG hydroxy methylated version and an all CpG unmethylated version) was used as a control to detemermine 5hmC scores.
Immunohistochemical Staining
Immunohistochemistry (IHC) was performed in cut sections of lung tumors using anti-MUC4 MAb clone 8G7 (MAb MUC4/8G7, the kind gift of Surinder K. Batra) [9] using the immunoperoxidase method. Antigen retrieval was performed using CC1 antigen retrieval buffer (pH 8.5, EDTA, 100°C, 30 minutes; Ventana Medical Systems, AZ, USA) for all sections. Following incubation in phosphate buffered saline, pH 7.4 (PBS) with 1% bovine serum albumin (BSA), sections were stained on a Benchmark XT automated slide stainer using a diaminobenzidine detection kit (UltraView DAB, Ventana Medical Systems). The control staining (normal mouse serum or PBS-BSA instead of the primary antibodies) showed no reaction.
RNA interference
TET1 knockdown was performed using MISSION® esiRNA human TET1 (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. MISSION® siRNA Universal Negative Control (Sigma-Aldrich, St. Louis, MO, USA) was used as a control. Briefly, A427 and NCI-H292 cells were seeded in 6-cm dishes. At 50% confluency cells were transfected with 13.6 nmol/l siRNA using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). After 48 h incubation, the cells were harvested.
Statistical Analysis
Data were analyzed using the “R” computing environment [37]. The normality of the data distribution was evaluated by the Kolmogorov-Smirnov test. An F test was performed to compare the variances of the two samples from normal populations. A non-parametric test of two-group difference was performed by the Mann-Whitney U test. A parametric test of two-group difference was performed by the Welch t-test (Unequal variance) or Student t-test (Equal variance). A Bartlett test was performed to compare the variances of multiple samples from normal populations. A nonparametric test of multi-group difference was performed by the Kruskal-Wallis one-way analysis of variance. A parametric test of multi-group difference was performed by the one-way analysis of variance (ANOVA). The correlation coefficient (R) was determined by the Pearson product-moment correlation coefficient. The multiple regression analysis was performed with the general linear model and goodness of fit was analyzed with coefficient of determination (R squared) values. The threshold points were determined by ROC curve analysis. A p-value <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Yukari Nishimura for her excellent technical assistance with immunohistochemistry.
ABBREVIATIONS
Mucins (MUC), immunohistochemistry (IHC), Ten-Eleven Translocation (TET), 5-methyl cytosine (5mC), 5-hydroxy methyl cytosine (5hmC), DNA methyltransferase (Dnmt), activation-induced deaminase (AIDCA), apolipoprotein B mRNA editing enzyme, catalytic polypeptide (APOBEC), glial cells missing (GCM).
FUNDING
This study was supported in part by a grant from Grants-in-Aid for Scientific Research on Scientific Research (C) 15K08466 to M. Higashi, Scientific Research (C) 15K11297 to T. Hamada and Young Scientists (B) 15K21247 to S. Yokoyama from the Ministry of Education, Science, Sports, Culture and Technology, Japan; by the Kodama Memorial Foundation, Japan (S. Yokoyama & M. Higashi). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CONFLICTS OF INTEREST
We have no conflict of interest to disclose concerning this study.

- 1. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012; 62: 220-41. [PubMed]
- 2. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65: 87-108. [PubMed]
- 3. (2016). SEER Cancer Statistics Review, 1975-2013, National Cancer Institute.: Bethesda, MD,), pp. based on November 2015 SEER data submission, posted to the SEER web site. [PubMed]
- 4. MUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cells. Cancer Res. 2008; 68: 2065-70. [PubMed] https://doi.org/10.1158/0008-5472.CAN-07-6041.
- 5. MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol Cancer Res. 2007; 5: 309-20. [PubMed]
- 6. Muc4/sialomucin complex, the intramembrane ErbB2 ligand, induces specific phosphorylation of ErbB2 and enhances expression of p27(kip), but does not activate mitogen-activated kinase or protein kinaseB/Akt pathways. Oncogene. 2002; 21: 752432. [PubMed]
- 7. A role for human MUC4 mucin gene, the ErbB2 ligand, as a target of TGF-beta in pancreatic carcinogenesis. Oncogene. 2004; 23: 5729-38. [PubMed]
- 8. Human MUC4 mucin induces ultra-structural changes and tumorigenicity in pancreatic cancer cells. Br J Cancer. 2007; 97: 345-57. [PubMed] https://doi.org/10.1038/sj.bjc.6603868.
- 9. Generation and characterization of anti-MUC4 monoclonal antibodies reactive with normal and cancer cells in humans. J Histochem Cytochem. 2004; 52: 253-61. [PubMed]
- 10. MUC4 activates HER2 signalling and enhances the motility of human ovarian cancer cells. Br J Cancer. 2008; 99: 520-6. [PubMed] https://doi.org/10.1038/sj.bjc.6604517.
- 11. Emerging roles of MUC4 in cancer: a novel target for diagnosis and therapy. Cancer Res. 2007; 67: 433-6. [PubMed]
- 12. MUC4 expression correlates with poor prognosis in small-sized lung adenocarcinoma. Lung Cancer. 2007; 55: 195-203. [PubMed]
- 13. Combined status of MUC1 mucin and surfactant apoprotein A expression can predict the outcome of patients with small-size lung adenocarcinoma. Histopathology. 2004; 44: 147-55. [PubMed]
- 14. Expression profiles of MUC1, MUC2, and MUC4 mucins in human neoplasms and their relationship with biological behavior. Proteomics. 2008; 8: 3329-41. [PubMed]
- 15. Epigenetic regulation of mucin genes in human cancers. Clin Epigenetics. 2011; 2: 85-96. [PubMed] https://doi.org/10.1007/s13148-011-0037-3.
- 16. Promoter CpG methylation in cancer cells contributes to the regulation of MUC4. Br J Cancer. 2009; 100: 344-51. [PubMed] https://doi.org/10.1038/sj.bjc.6604845.
- 17. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008; 68: 270816. [PubMed]
- 18. Diagnosis of pancreatic neoplasms using a novel method of DNA methylation analysis of mucin expression in pancreatic juice. PLoS One. 2014; 9: e93760. [PubMed] https://doi.org/10.1371/journal.pone.0093760.
- 19. The application of methylation specific electrophoresis (MSE) to DNA methylation analysis of the 5’ CpG island of mucin in cancer cells. BMC Cancer. 2012; 12: 67. [PubMed] https://doi.org/10.1186/1471-2407-12-67.
- 20. Aberrant methylation of MUC1 and MUC4 promoters are potential prognostic biomarkers for pancreatic ductal adenocarcinomas. Oncotarget. 2016; 7: 42553-65. [PubMed] https://doi.org/10.18632/oncotarget.9924.
- 21. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum Mol Genet. 2001; 10: 2917-31. [PubMed]
- 22. Genome-wide survey reveals dynamic widespread tissue-specific changes in DNA methylation during development. BMC Genomics. 2011; 12: 231. [PubMed] https://doi.org/10.1186/1471-2164-12-231.
- 23. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007; 447: 425-32. [PubMed]
- 24. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011; 42: 451-64. [PubMed] https://doi.org/10.1016/j.molcel.2011.04.005.
- 25. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992; 69: 915-26. [PubMed]
- 26. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99: 247-57. [PubMed]
- 27. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333: 1300-3. [PubMed] https://doi.org/10.1126/science.1210597.
- 28. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011; 145: 423-34. [PubMed] https://doi.org/10.1016/j.cell.2011.03.022.
- 29. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012; 28: 164-74. [PubMed]
- 30. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011; 333: 1303-7. [PubMed] https://doi.org/10.1126/science.1210944.
- 31. Mammalian Gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat Neurosci. 2011; 14: 957-64. [PubMed]
- 32. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011; 2: 627-37. [PubMed] https://doi.org/10.18632/oncotarget.316.
- 33. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012; 150: 1135-46. [PubMed] https://doi.org/10.1016/j.cell.2012.07.033.
- 34. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011; 19: 17-30. [PubMed] https://doi.org/10.1016/j.ccr.2010.12.014.
- 35. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013; 32: 663-9. [PubMed] https://doi.org/10.1038/onc.2012.67.
- 36. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012; 149: 1368-80. [PubMed] https://doi.org/10.1016/j.cell.2012.04.027.
- 37. R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996; 5: 16.
Last Modified: 2017-06-22 00:28:03 EDT
PII: 139
