Genes & Cancer
Abstract | PDF | Full Text | Supplementary Materials | Supplementary Materials
https://doi.org/10.18632/genesandcancer.164
Dysregulation of AKT3 along with a small panel of mRNAs stratifies high-grade serous ovarian cancer from both normal epithelia and benign tumor tissues
Pourya Naderi Yeganeh1, Christine Richardson2, Zahra Bahrani-Mostafavi3, David L. Tait4, M. Taghi Mostafavi1
1 College of Computing and Informatics, University of North Carolina at Charlotte, Charlotte, NC, USA
2 Department of Biological Sciences, University of North Carolina at Charlotte, Charlotte, NC, USA
3 College of Health and Human Services, University of North Carolina at Charlotte, Charlotte, NC, USA
4 Division of Gynecological Oncology and Obstetrics, Levine Cancer Institute, Carolinas Medical Center, Charlotte, NC, USA
Correspondence to: M. Taghi Mostafavi, email: [email protected]
Keywords: ovarian cancer, AKT3, genomic analysis, high-grade serous ovarian cancer, biomarker discovery
Received: September 20, 2017
Accepted: December 01, 2017
Published: December 29, 2017
ABSTRACT
Screening methods of High-Grade Serous Ovarian Cancer (HGSOC) lack specificity and sensitivity, partly due to benign tumors producing false-positive findings. We utilized a differential expression analysis pipeline on malignant tumor (MT) and normal epithelial (NE) samples, and also filtered the results to discriminate between MT and benign tumor (BT). We report that a panel of 26 dysregulated genes stratifies MT from both BT and NE. We further validated our findings by utilizing unsupervised clustering methods on two independent datasets. We show that the 26-genes panel completely distinguishes HGSOC from NE, and produces a more accurate classification between HGSOC and BT. Pathway analysis reveals that AKT3 is of particular significance, because of its high fold change and appearance in the majority of the dysregulated pathways. mRNA patterns of AKT3 suggest essential connections with tumor growth and metastasis, as well as a strong biomarker potential when used with 3 other genes (PTTG1, MND1, CENPF). Our results show that dysregulation of the 26-mRNA signature panel provides an evidence of malignancy and contribute to the design of a high specificity biomarker panel for detection of HGSOC, potentially in an early more curable stage.
INTRODUCTION
Ovarian cancer is the leading cause of death among gynecological cancers and early diagnosis is one of the key challenges in reducing the mortality rate [1-3]. Early detection strategies of ovarian cancer lack required specificity and sensitivity, leading to diagnosis in advanced and more lethal stages in 75% of patients [4, 5]. For example, elevations of the biomarker Cancer Antigen 125 (CA125) produce a relatively small true positive rate for early stage ovarian cancers, while the false positive rates are high for patients with benign tumors and non-cancerous subjects [5, 6]. Major clinical trials -- Prostate, Lung, Colon, and Ovarian (PLCO) screening trials, and UK Collaborative Trial of Ovarian Cancer Screening (UKTOCS) – indicate that the current screening strategies do not contribute to the reduction of mortality rates [7, 8]. Not surprisingly, the design and discovery of credible biomarker panels for detection of ovarian cancer has emerged as a formidable task.
To date, genome-scale comparative studies have considerably contributed to the discovery of new markers, therapeutic targets, cancer subtypes, and origins of ovarian cancer. [9-13]. The utilization of genomic scale analyses has also substantially benefited the characterization of malignant tumors [14, 15]. However, many of the genetic and pathological features associated with cancer also occur in benign tumors, including some of the hallmarks of cancer, such as evading growth suppressors and resisting cell death [14-16]. The molecular and pathological resemblance between malignant and non-malignant tumors is a barrier in the design of cancer-focused detection and treatment approaches [16]. Given these challenges, the identification of malignancy-specific molecular signatures plays a critical role in the disease diagnosis and management.
This study focuses on identification of a molecular signature panel in High-Grade Serous Ovarian Cancer (HGSOC) which constitutes up to 80% of the cases of ovarian cancer [17]. We focus on the discovery of a small signature panel of mRNA markers that can differentiate between malignant and benign tumors. We hypothesize that comparative mRNA analysis of ovarian tissues would reveal critical signatures of malignancy and tumor progression in HGSOC. To probe our hypothesis, we investigated the gene expression profiles of distinct ovarian tissues by utilizing Laser Capture Microdissection (LCM) for high sample purity [18]. We analyzed the mRNA expression levels of normal ovarian epithelia (NE), benign ovarian tumors (BT), and HGSOC malignant tumors (MT). Analyses were performed to capture the differential expressions patterns, similar to Bowen et al. [9] who utilized a related design and identified ovarian surface epithelial markers in the cancer tumors. Our experimental design uniquely included BT samples which allowed for a more comprehensive molecular characterization of the different pathological states [16]. Our results identify a small signature mRNA panel of 26 dysregulated genes that stratifies MT from both BT and NE. Validity of the identified panel was verified by using classification algorithms on two publicly available independent datasets (GSE9899 and GSE14407) [9, 12]. To determine the functional properties of the dysregulated genes in the active programs of HGSOC, we compared results from pathway enrichments and the mRNA dysregulation between MT and BT.
RESULTS
Differential mRNA expression analysis of normal epithelia versus malignant tumors:
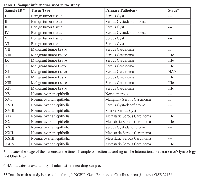
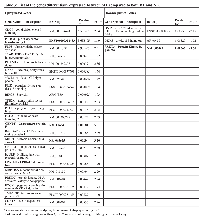
Analysis of MT and NE samples (Table 1) detects 216 significantly differentially expressed genes, 100 of which are upregulated and 116 are downregulated (supplementary table). Table 2 details the 20 upregulated genes with greatest fold changes as well as the 20 downregulated genes with greatest fold changes. The analysis detects dysregulation of genes previously associated with HGSOC – such as Paired box 8 (PAX8), Paternally expressed 3 (PEG3), Survivin (BIRC5), Meis homeobox 1 (MEIS1), and Homeobox C6 (HOXC6) - as well as unreported ones, such as Meiotic nuclear divisions 1 (MND1), and Protein Kinase B, Gamma (AKT3), (Table 2 and supplementary table) [19-21].
Dysregulated mRNA expressions between benign and malignant tumors:
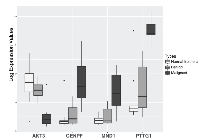
At the initial step of the analysis pipeline, 216 dysregulated genes were found between MT and NE. The dysregulated genes were then tested in another specific comparison pipeline. Of the 216 dysregulated genes 26 were detected as differentially expressed between MT and BT (Table 3). Among the 26 genes, AKT3, Human Securin encoding gene (PTTG1), Centromere protein F (CENPF), and MND1 exhibit fold change magnitudes of greater than 3 (Figure 1, Table 3). In MT vs. NE setting, AKT3 is downregulated by 4.68 folds (p-value < 2.99 E-04), and PTTG1, CENPF, and MND1 are all upregulated by 28.8 fold (p-value < 1.96 E-06), 5.9 fold (p-value < 2.1 E-04), and 3.9 fold (p-value < 1.29 E-04), respectively. In MT vs. BT setting, the mRNA dysregulation of these four genes (AKT3, PTTG1, CENPF, and MND1) yield a similar statistically significant pattern (Table 3).
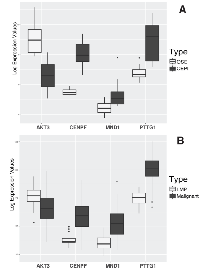
Cross-examination of expression of the set of four genes provides additional evidence for their dysregulation in HGSOC. Two independent and publicly available datasets by Tothill et al. (GSE9899) and Bowen et al. (GSE14407) (Figure 2) [9, 12] were utilized. In malignant tumors compared to Low Malignant Potential (LMP) tumors in GSE9899 dataset, AKT3 is downregulated by 2.1 folds (p-value < 0.0012); PTTG1, MND1, and CENPF are upregulated with 5.9 (p-value < 2.83 E-19), 4.12 (p-value 9.9 E-11), and 3.17 folds (p-value < 1.67E-08), respectively. The dysregulation patterns persist in LCM-collected normal epithelia and malignant epithelial ovarian tissues (from GSE14407). AKT3 is downregulated by 5.7 folds (p-value < 4.11 E-06); PTTG1, MND1, and CENPF are upregulated with 4.2 (p-value < 3.52 E-05), 1.86 (p-value < 0.008), and 5.78 folds (p-value < 1.87 E-09), respectively. Although the experimental design and samples are different in each study, the displayed results validate the findings. The test datasets contain control samples from normal and benign ovarian tissues, and provide a suitable sample composition for validating our findings.
Enrichment analysis results of the 26 differentially expressed genes between MT and BT detects the terms cell cycle, regulation of mitotic cell cycle phase transition, regulation of cell cycle phase transition, E2F cell cycle related transcription factors, and G2/M checkpoint in hallmarks. Common biological knowledge assumes that the malignant cells exhibit attributes indicating invasion and rapid cell growth compared to benign cells. PTTG1, CENPF, BIRC5, Enhancer of zeste homolog 2 (EZH2), Rac GTPase activating protein 1 (RACGAP1), and cell division cycle 7 homolog (CDC7) are associated with G2M checkpoint (p-value < 3.16 E-6). Similarly, BIRC5, EZH2, RACGAP1, PTTG1, and protein kinase, DNA-activated, catalytic polypeptide (PRKDC) are associated with E2F transcription factor cell cycle-related targets (p-value < 1.06 E-4).
A panel of 26 dysregulated genes distinguishes HGSOC tumors from normal and benign ovarian samples:
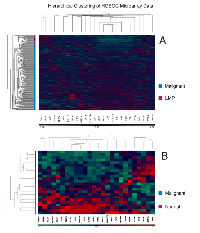
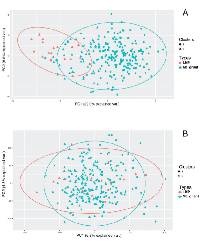
The two datasets (GSE14407 and GSE9899) were used to investigate the utility of the 26 dysregulated genes (Table 3) as a mRNA panel to stratify malignant samples from benign and normal samples. Unsupervised hierarchical clustering of the mRNA panel on GSE9899 shows that LMP samples are clustered close together and separated from malignant samples (Figure 3A). Using a similar procedure on GSE14407 shows a clear distinction between the normal epithelial samples and malignant samples (Figure 3B).
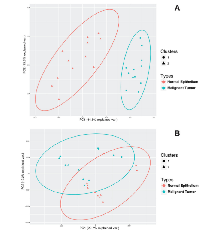
Principal Component Analysis (PCA), followed by k-means clustering, was used to further assess the utility of the mRNA panel. PCA of GSE14407 samples shows that the use of the 26-genes mRNA panel is sufficient to stratify normal epithelial tissues from HGSOC (Table 4). The unsupervised k-means clustering separates the tissue types into two distinct groups with no overlap between their 95% confidence ellipses, when only using the two first principal components (Figure 4). In contrast, a similar procedure fails to distinguish between the tissue types when the mRNA signature panel is not used for PCA; and finds an overlap between the 95% confidence ellipses. When using the mRNA panel, the first two principal components explain more than 59.6% of the variance in the data, up from 48.9% when using all the genes for PCA. PCA followed by k-means clustering on GSE9899 dataset shows that using the dysregulated mRNA panel produces a stronger classification compared to when using all genes (Figure 5). k-means clustering classifies LMP samples into the same group and associates a larger fraction of the malignant tumors to another group when using the 26-genes mRNA panel as a base. In contrast, using all genes’ transcripts fails to discriminate between the two groups and distributes the tissue types in both of the clusters (Table 4). Also, using the mRNA panel increases the variance in data explained by PCA. The first two principal components explain 35.9% of the variance when using the 26-panel, up from 11.1% when using no panel (Figure 5).
Pathways and Interactions analysis:
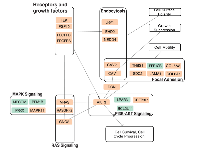
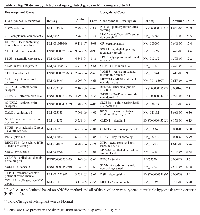
Pathway enrichment analysis using the 26-genes signature panel (Table 3) identified 10 KEGG annotated human pathways. The results show dysregulation of critical cellular mechanisms and signaling pathways. The dysregulated pathways include endocytosis, focal adhesion, MAPK signaling, PI3K-AKT signaling, and RAS signaling (Table 5). P-values of enrichment analysis are calculated using chi-squared test. The analysis was carried through Partek Software Package and KEGG pathway database. The overlaps of the identified pathways are summarized as a network of dysregulation, with some additional interactions included from STRING database. Figure 6 displays the network and illustrates how each module participates in the enriched pathways and biological functions. Connections of elements are either direct molecular interactions or mediated interactions. Some of the dysregulated genes are shared between multiple pathways. For instance, Caveolins (CAV1, CAV2) are shared between endocytosis and focal adhesion. AKT3, CAV1, CAV2, Platelet Derived Growth Factor Receptor A (PDGFRA), and PDGFRB are shared between four pathways. Those include focal adhesion MAPK, PI3K-AKT, and RAS signaling (Figure 6).
This analysis demonstrates that AKT3 and PDGFRA appear as the central elements in terms of the position in the network, overlaps between the dysregulated pathways, and the fold change magnitude. The results show mRNA downregulation of PDGFRA in MT. Lower mRNA expression of AKT3 are observable in MT compared to BT and NE (Table 2 and Table 3). AKT3 is involved in 8 of the 10 identified pathways – Dopaminergic synapse, Rap1 signaling, Focal adhesion, MAPK, Melanoma, Proteoglycans in cancer, PI3K-AKT, and Ras signaling (Table 5). PDGFRA is associated with 7 of the pathways (supplementary material). Two of the dysregulated genes in MT compared to BT, AKT3 and BCL11, are also involved in the extracted network.
DISCUSSION
We conducted a comparative analysis of mRNA expression using laser capture micro-dissected ovarian tissues from NE, BT, and MT. Differential expression analysis identifies dysregulated genes that stratify MT from both NE and BT (Tables 2 and 3). The set of four genes AKT3, PTTG1, CENPF, and MND1 are the most significant, based on fold changes, among the dysregulated genes in MT compared to BT and NE (Figure 1). To verify this observation, we utilized two independent publicly available datasets (GSE14407 and GSE9899) (Figure 2). Cross-examination results show statistically significant evidence of consistent dysregulations of four genes (Figure 2). The distinguishing expression patterns of these genes, between the different pathological states, suggest potential roles as indicators of HGSOC malignancy. Prior studies support the roles of some of these genes in ovarian cancer. PTTG1 is a therapeutic target in ovarian cancer and is associated with functions including DNA repair, angiogenesis, and cell development [22, 23]. CENPF expression has been associated with cell cycle progression, and malignancy through FOXM1 [20, 24]. To our knowledge, AKT3 downregulation and MND1 upregulation have not been previously reported in human HGSOC.
While dysregulations of the set of fours genes are significant between MT vs BT and NE, there are also some detectable dysregulation between BT and NE (Figures 1 and 2B). As the dysregulations indicate the tumor presence, their intensity can determine whether the tumors are malignant or not. Moderate dysregulation of BT as compared to NE is supported by the fact that benign tumors exhibit functional similarities with malignant tumors, including evading growth suppression and resisting cell death [14, 16]. However, malignant tumors are expected to exhibit more extreme dysfunctions as well as other unique characteristics such as metastasis [14]. In our study, enrichment analysis results of the 26 dysregulated signature gene panel (MT vs BT and NE) identified G2M checkpoint and E2F transcription factor cell cycle-related targets, which are related with P53 activities [25, 26]. These results are supported by the reports of HGSOC displaying P53 mutations in more than 70% of cases [1, 27], compared to dramatically lower rates in low malignant potential tumors [28].
The classification results display the utility of the 26-gene signature panel to distinguish between MT and both BT and NE when the signals are combined (Figure 4 and 5, Table 4). PCA combines mRNA signals and represent the microarray samples in a lower dimensional space, which allows for an interpretable feature selection. k-means clustering shows that the 26-genes panel stratify the PCA-projected samples, based on their respective tissue types (Figures 4A and 5A). In a similar procedure, without filtering the input mRNA signals, the classification algorithm loses its stratification power (Table 4). This observation indicates that the comparative analysis of LCM samples generates a focused and meaningful outline of the genetic dysregulations necessary for identification of HGSOC. As HGSOC is known to encompass a diverse genetic profile, the results show how the use of mRNA panel boosts the classification quality and addresses the issue of genetic heterogeneity. This also indicates that the signature mRNA panel can contribute to the design of future collective biomarkers as alternative to single-molecule biomarkers.
Pathway enrichment analysis identifies functional associations of the dysregulated genes, and shows major overlaps among the ten enriched KEGG pathways (Table 5, Figure 6). In particular, AKT3 appears in 8 and PDGFRA appears in 7 of the 10 pathways. PDGFRA is connected to AKT3 activity and regulations in participates in cancer related mechanisms [29]. PDGFRA alteration is associated with ovarian cancer but not frequently mutated [30, 31]. This suggests that the interconnections between AKT3 and PDGFRA are potential for future studies. The extracted network of pathway overlaps shows associations of HGSOC dysregulation with several critical cancer mechanisms, including cell survival, cell motility, and tumor growth. Additional evidence from the literature indicates that endocytosis and focal adhesion components of this network are involved in Epithelial-Mesenchymal Transition (EMT), and cell motility [32-34].
Based on the dysregulation patterns and functional associations, our results indicate that AKT3 is a potential central marker of tumor presence and malignancy. AKT3 is one of the 3 major protein kinase B isoforms, and exhibits high levels of homology to the other members of the AKT family (AKT1 and AKT2), particularly in the highly conserved phosphorylation sites [35]. AKTs are frequently dysregulated in ovarian cancer, and participate in various cancer associated activities, including proliferation, migration, survival, and apoptosis [35-41]. While AKT3 is relatively understudied compared to the other members of AKT family, its downregulation is to some extent contrary to the early literature [39, 42, 43]. Earlier studies reported upregulation of AKT molecules in breast, prostate, and ovarian cancers [35, 39, 42, 43]. However, our results show that AKT3 is downregulated, which is also supported by more recent studies. Phung et al. showed that AKT3 inhibits growth in vascular tumors and displays declined expression [40]. Grottke et al. also showed that AKT3 downregulation results in elevated migration and metastasis in breast cancer [44]. Based on validation in independent public datasets (GSE9899 and GSE14407), we conclude that AKT3 downregulation in HGSOC implies opposing patterns compared to reports of AKT1 and AKT2 [20]. In addition, we identify AKT3 as a potential central marker of tumor malignancy, as its levels in BT are slightly lower than in NE, and significantly further decreased in MT compared to BT (Figure 2). Nakatani et al. reported that AKT3 enzyme activity levels correlate with its upregulated mRNA levels in breast cancer and prostate cancer [39]. This finding, along with our results, suggests that AKT3 mRNA downregulation could reduce its protein phosphorylation activity and potentially can be used to establish a strong HGSOC biomarker. In addition, AKT3 downregulations might contribute to tumor growth, metastasis, and cell migration in HGSOC [40, 44]. We conclude that further study of AKT3 activities in ovarian cancer will lead to new insights of the signaling activities, especially those related to the PI3K-AKT pathway.
The results presented in this article display how the comparative investigation of mRNAs provides signatures for identifying HGSOC relative to both normal epithelia and benign tumors. Coupled with pathway analysis, we determine functional dysregulations associated with these signatures. The activities of the dysregulated genes and their causal relationships need to be further investigated using other molecular aspects such as protein assays, DNA methylation, and phosphorylation. The significant changes in mRNA levels of the 26-gene signature panel indicate their potential as biomarkers for HGSOC, due to their specificity to malignant tissues. We showed how the dysregulated genes could be used as molecular signatures to stratify HGSOC (Figure 2). These molecular signatures can potentially be used to enhance early detection for HGSOC and ultimately contribute to managing mortality rates.
MATERIALS AND METHODS
Our initial in-house data set consists of 24 cell specific samples processed through LCM, 10 normal epithelial tissues (NE), 8 malignant tumor tissues (MT), and 6 benign tumor tissues (BT) (Table 1). In addition, we utilized two related and independently collected datasets for verification of results in larger datasets, GSE14407 and GSE9899. GSE14407 consists of 12 normal ovarian epithelial tissues and 12 ovarian malignant tumor tissues. GSE9899 includes 18 low malignant potential samples and 246 HGSOC samples. Our in-house data is publicly available through NCBI Gene Expression Omnibus using reference code GSE 29156. Non-tumorous ovarian epithelial tissues, and tumor tissues from benign and malignant serous carcinoma samples were analyzed with GeneChip Affymetrix Human Exon 1.0 ST arrays. Gene expressions were subsequently subjected to multiple statistical procedures for production of accurate and quality results.
Tissue collection
Ovarian specimens were obtained, under IRB approved guidelines at Carolinas Medical Center (CMC), during surgery for ovarian cancer or other gynecologic conditions. The samples were classified as normal ovary, benign tumors, or malignant tumors. Malignant tissues were from patients with serous carcinoma at stage II or III, grade 3 according to pathological diagnosis assigned by pathologists at CMC using the World Health Organization criteria for ovarian tumors (Table 1). Tissue samples were placed in a standard sized cryomold (Sakura Finetek USA, Inc., Torrance, CA), covered with Optimal Cutting Temperature (OCT) compound (Sakura Finetek USA, Inc., Torrance, CA), frozen and stored at -80°C [19].
Laser Capture Microdissection (LCM)
OCT embedded samples were serially sectioned into 8μm sections and prepared for LCM using the HistoGene LCM Frozen Section Staining kit (Applied Biosystems, Life Technologies, Co., Carlsbad, CA) according to the manufacturer’s protocols. The stained sections were immediately micro-dissected by an Arcturus® PixCell® IIe LCM (Molecular Devices, LLC, Sunnyvale, CA). Normal epithelium and tumor cells were separately collected from appropriate sections. RNA quality was determined using the residual slide materials [19].
cDNA Synthesis and Amplification
Single primer isothermal amplified (SPIA) complimentary DNA (cDNA) was generated and amplified using the Whole Transcriptome WT-Ovation Pico RNA Amplification System Kit (NuGEN Technologies Inc., San Carlos, CA), and was used for microarray sample preparation.
Exon Microarray and Sample Hybridization
3 μg of SPIA amplified cDNA was used to do the sense–strand cDNA (ST-cDNA) conversion using the WT-Ovation Exon Module (NuGEN Technologies Inc., San Carlos, CA). 5 μg ST-cDNA was fragmented and labeled with FL-Ovation cDNA Biotin Module V2 kit (NuGEN Technologies Inc., San Carlos, CA) and hybridized using Affymetrix Human Exon 1.0 ST arrays (Affymetrix, Inc., Santa Clara, CA). Microarray hybridization and scanning was performed using GeneChip Hybridization Oven 640, GeneChip Fluidics Station 450, and GeneChip Scanner 3000 7G with Autoloader respectively (Affymetrix, Inc., Santa Clara, CA) [19].
Statistical Analysis
The samples were analyzed using Partek® Genomics Suite® (Partek Inc., St. Louis, MO). Microarray transcripts were normalized using GCRMA method. Exons were summarized to genes expressions using winsorized mean (below 10.0% and above 90.0%) and Tukey’s biweight one-step. Differential expression p-values were calculated using Analysis of Variance (ANOVA). Unannotated genes were removed and transcripts passed as significant if their False Discovery Rate (FDR) criteria was less than 0.05 (FDR<0.05) (Table 2).
Differentially expressed genes between MT and NE were analyzed to determine differentially expressed genes between BT and MT (FDR < 0.05) (Table 3).
Validation and confirmation of results
We used two publicly available datasets to verify the results. First, we utilized an experimentally similar datasets from Bowen and colleagues which includes 12 normal ovarian epithelial tissues and 12 ovarian tumor tissues each separated using LCM [9]. The samples were obtained through NCBI GEO portal from GSE14407 dataset. Second, we used whole tissue samples by Tothill and colleagues (GSE9899); with samples limited to Low Malignant Potential (LMP) and ovarian serous carcinoma malignant tumors [12]. The samples of GSE9899 that we used included 18 LMBs and 246 malignant tumors. We used hierarchical clustering algorithm using average linkage and Euclidian distance to cluster the samples from GSE14407 and GSE9899. The hierarchical clustering procedure was done using Partek® Genomics Suite®. We used Principal Component Analysis (PCA) to evaluate the utility of dysregulated genes between MT and BT compared to all gene transcripts in distinguishing between the sample types (Figures 4 and 5). For each test dataset, we separately applied PCA to all annotated genes and the panel of dysregulated genes. PCA was calculated based on the covariance matrix of gene expressions and the data points were shifted to the mean of zero. For further evaluation, we used k-means algorithm for unsupervised clustering of samples into two groups after applying PCA. For each dataset, the results of k-means were calculated based on whether PCA was done on the dysregulated genes between MT and BT or not. PCA results were projected on the first two principle components. Cross validation of results were done using R statistical analysis package [45]. Moreover, the statistical significance of the dysregulated gene between MT and BT with fold change magnitude more than 3 were evaluated in the test datasets (Figure 2).
Enrichment Analysis
Differentially expressed genes in NE vs MT were further analyzed to investigate the underlying functional dysregulations. Overrepresentation analysis of KEGG annotated pathways was performed using Partek® Genomics Suite®. Enrichment score of each pathway was calculated based on Chi-squared test of differentially expressed gene in pathway, relative to the total number of the annotated genes of pathway (Table 5). P-values of Chi-squared test were subjected to multiple hypothesis testing criteria (FDR < 0.05). Additional interactions were included by manual examination of the Protein-Protein Interactions (PPI) network of Homo Sapiens. PPIs were obtained using STRING-DB online database [46]. The interactions were included if they had medium or higher confidence based on data input of STRING-DB criteria [46]. Gene Ontology (GO) enrichments and related annotated enrichments were investigated based on MSigDB database by Broad Institute [47]. We used the same statistical criteria (FDR < 0.05) as the cut off for reporting enriched terms.
Abbreviations
BT: Benign Tumors, NE: Normal Epithelia, MT: Malignant Tumors, LCM: Laser Capture Microdissection, LMP: Low Malignant Potential, HGSOC: High-Grade Serous Ovarian Cancer, PCA: Principal Component Analysis.
AUTHORS’ CONTRIBUTIONS
PNY: Conception and design, development of methodology, analysis and interpretation of data, writing, review and/or revision of the manuscript.
CR: Writing, review and/or revision of the manuscript, analysis and interpretation of data.
ZBM: Administrative, technical, or material support, development of methodology, conception and design.
DLT: Acquisition of data, review and/or revision of the manuscript, analysis and interpretation of data.
MTM: Study supervision, writing, review and/or revision of the manuscript, administrative, technical, or material support.
ACKNOWLEDGEMENTS
We would like to thank the National Cancer Institute for making their publicly available data. Also, we would like to thank extensive and continuous support from Carolinas Healthcare System and the College of Computing and Informatics at UNC Charlotte. We would like to express our gratitude to those who made this study possible by donating their samples.
CONFLICTS OF INTEREST
Authors declare no potential conflicts of interest.
Ethics approval and consent to participate
Carolinas HealthCare System IRB# 05-05-16B.
Availability of data and materials
The original data used in this study is published and also available in Gene Expression Omnibus. The accession code is GSE 29156.
- 1. Ovarian cancer: pathology, biology, and disease models. Frontiers in bioscience: a journal and virtual library. 2009; 14: 2089. [PubMed]
- 2. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016; 66: 7-30. [PubMed]
- 3. SEER Cancer Statistics Review, 1975-2013, National Cancer Institute. Bethesda, MD. http://seercancergov/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site. [PubMed]
- 4. Beyond CA125: the coming of age of ovarian cancer biomarkers. Biomarkers in medicine. 2009; 3: 275-88. [PubMed] https://doi.org/10.2217/bmm.09.21.
- 5. Biomarkers for early detection of ovarian cancer. Women’s health. 2013; 9: 17187. [PubMed]
- 6. Detection and monitoring of ovarian cancer. Clinica Chimica Acta. 2013; 415: 341-5. [PubMed]
- 7. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening randomized controlled trial. Jama. 2011; 305: 2295-303. [PubMed]
- 8. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. The Lancet. 2016; 387: 945-56. [PubMed] https://doi.org/10.1016/S0140-6736(15)01224-6.
- 9. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC medical genomics. 2009; 2: 1. [PubMed] https://doi.org/10.1186/1755-8794-2-71.
- 10. Predicting biomarkers for ovarian cancer using gene-expression microarrays. British journal of cancer. 2004; 90: 686-92. [PubMed]
- 11. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. International Journal of Cancer. 2004; 112: 1425. [PubMed] https://doi.org/10.1038/sj.bjc.6601603.
- 12. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clinical Cancer Research. 2008; 14: 5198-208. [PubMed]
- 13. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proceedings of the National Academy of Sciences. 2001; 98: 1176-81. [PubMed] https://doi.org/10.1073/pnas.98.3.1176.
- 14. Hallmarks of Cancer: The Next Generation. Cell. 2011; 144: 646-74. [PubMed]
- 15. The dominance of the microenvironment in breast and ovarian cancer. Seminars in Cancer Biology. 2002; 12: 97-104. [PubMed] https://doi.org/10.1006/scbi.2001.0417.
- 16. What are the hallmarks of cancer? Nature Reviews Cancer. 2010; 10: 232-3 [PubMed]
- 17. Rethinking ovarian cancer II: reducing mortality from highgrade serous ovarian cancer. Nature Reviews Cancer. 2015; 15: 668-79. [PubMed] https://doi.org/10.1038/nrc4019.
- 18. Lasercapture microdissection. Nature protocols. 2006; 1: 586603. [PubMed]
- 19. Downregulation of HOXC6 in serous ovarian cancer. Cancer investigation. 2015; 33: 303-11. [PubMed]
- 20. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474: 609-15. [PubMed] https://doi.org/10.1038/nature10166.
- 21. Emerging roles for PAX8 in ovarian cancer and endosalpingeal development. Gynecologic oncology. 2007; 104: 331-7. [PubMed]
- 22. PTTG: an important target gene for ovarian cancer therapy. Journal of ovarian research. 2008; 1: 1. [PubMed] https://doi.org/10.1186/1757-2215-1-6.
- 23. Molecular cloning of pituitary tumor transforming gene 1 from ovarian tumors and its expression in tumors. Cancer letters. 2001; 163: 1319. [PubMed]
- 24. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer cell. 2014; 25: 638-51. [PubMed] https://doi.org/10.1016/j.ccr.2014.03.017.
- 25. p53 and E2f: partners in life and death. Nature Reviews Cancer. 2009; 9: 738-48. [PubMed]
- 26. Regulation of the G2/M transition by p53. Oncogene. 2001; 20: 1803-15. [PubMed]
- 27. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. The Journal of pathology. 2010; 221: 49-56. [PubMed] https://doi.org/10.1002/path.2696.
- 28. A review of p53 expression and mutation in human benign, low malignant potential, and invasive epithelial ovarian tumors. Cancer. 2003; 97: 389-404. [PubMed]
- 29. Role of platelet-derived growth factors in physiology and medicine. Genes & development. 2008; 22: 1276-312. [PubMed] https://doi.org/10.1101/gad.1653708.
- 30. Genetic alterations and protein expression of KIT and PDGFRA in serous ovarian carcinoma. British journal of cancer. 2004; 91: 2048-55. [PubMed] https://doi.org/10.1038/sj.bjc.6602252.
- 31. Expression and prognostic significance of platelet-derived growth factor and its receptors in epithelial ovarian neoplasms. Cancer Research. 1993; 53: 4550-4. [PubMed]
- 32. Derailed endocytosis: an emerging feature of cancer. Nature Reviews Cancer. 2008; 8: 835-50. [PubMed]
- 33. Endocytosis and cancer. Cold Spring Harbor perspectives in biology. 2013; 5: a016949. [PubMed] https://doi.org/10.1101/cshperspect.a016949.
- 34. Biological significance of focal adhesion kinase in ovarian cancer: role in migration and invasion. The American journal of pathology. 2004; 165: 1087-95. [PubMed] https://doi.org/10.1016/S0002-9440(10)63370-6.
- 35. Frequent activation of AKT2 and induction of apoptosis by inhibition of phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer. Oncogene. 2000; 19: 2324-30. [PubMed]
- 36. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial–mesenchymal transition. The Journal of cell biology. 2005; 171: 1023-34. [PubMed] https://doi.org/10.1083/jcb.200505087.
- 37. Protein kinase Bα/Akt1 regulates placental development and fetal growth. Journal of Biological Chemistry. 2003; 278: 32124-31. [PubMed]
- 38. Targeting Akt3 signaling in triple-negative breast cancer. Cancer research. 2014; 74: 964-73. [PubMed] https://doi.org/10.1158/0008-5472.CAN-13-2175.
- 39. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. Journal of Biological Chemistry. 1999; 274: 21528-32. [PubMed]
- 40. Akt1 and Akt3 Exert Opposing Roles in the Regulation of Vascular Tumor Growth. Cancer Research. 2015; 75: 40-50. [PubMed]
- 41. PIK3CA is implicated as an oncogene in ovarian cancer. Nature genetics. 1999; 21: 99-102. [PubMed]
- 42. A specific role for AKT3 in the genesis of ovarian cancer through modulation of G2-M phase transition. Cancer Research. 2006; 66: 11718-25. [PubMed]
- 43. Akt3 controls vascular endothelial growth factor secretion and angiogenesis in ovarian cancer cells. International journal of cancer. 2012; 130: 532-43. [PubMed] https://doi.org/10.1002/ijc.26010.
- 44. Downregulation of AKT3 Increases Migration and Metastasis in Triple Negative Breast Cancer Cells by Upregulating S100A4. PloS one. 2016; 11: e0146370. [PubMed] https://doi.org/10.1371/journal.pone.0146370.
- 45. R: A language and environment for statistical computing. R Foundation for Statistical Computing Vienna, Austria. 2017; doi. [PubMed]
- 46. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids research. 2014; doi: gku1003. [PubMed] https://doi.org/10.1093/nar/gku1003.
- 47. Gene set enrichment analysis: a knowledgebased approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005; 102: 15545-50. [PubMed] https://doi.org/10.1073/pnas.0506580102.
Last Modified: 2017-12-30 19:42:03 EST
PII: 164
