Genes & Cancer
Alterations in TGF-β signaling leads to high HMGA2 levels potentially through modulation of PJA1/SMAD3 in HCC cells
Kazufumi Ohshiro1, Jian Chen2, Jigisha Srivastav3, Lopa Mishra1,4 and Bibhuti Mishra1
1 Department of Surgery, Center for Translational Medicine, George
Washington University, Washington DC, USA
2 Department of Gastroenterology, Hepatology,
and Nutrition, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
3
University of Toledo College of Medicine, Toledo, OH, USA
4 Department of
Gastroenterology and Hepatology, VA Medical Center, Washington DC, USA
Correspondence to: Lopa Mishra, email: [email protected]
Keywords: TGF-β pathway, HMGA2, PJA1, hepatocellular carcinoma
Received: October 07, 2019
Accepted: January 06, 2020
Published: January 22, 2020
Copyright: © 2020 Ohshiro et al. This is an open-access article distributed under the terms of the Creative Commons Attribution Li-cense 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Recently, we observed that the TGF-β pathway is altered in 39% of HCCs. The alterations are correlated with a raised HMGA2 level. Therefore, we compared genetic alterations of HMGA2 and 43 TGF-β pathway core genes in HCC patients from TCGA database. Genetic alterations of 15 genes, including INHBE, INHBC, GDF11, ACVRL and TGFB2 out of 43 core genes, highly-moderately matched that of HMGA2. Co-occurrences of mutation amplification, gains, deletions and high/low mRNA of HMGA2 with those of the core genes were highly significant in INHBE, INHBC, ACVR1B, ACVRL and GDF11. Mass spectrometry studies revealed that HMGA2 interacted with an E3 ligase, PJA1, and that this interaction is enhanced by TGF-β treatment in the nuclear of HCC cells. Co-localization of nuclear PJA1 and HMGA2 in HCC cells increased upon TGF-β treatment. Raised HMGA2 levels that occur with alterations in the TGF-β signaling pathway may reflect an altered activity of E3 ligases, such as PJA1, and potentially contribute to the tumor-promoting roles of TGF-β signaling. Here, we report that the co-occurrence of genetic alterations in HMGA2 and TGF-β pathway core genes is implicated in HCC progression, and propose that HMGA2 and PJA1 may be potential novel targets in dysfunctional TGF-β signaling in HCC.
INTRODUCTION
The incidence of HCC in the U.S. has increased 3-4 fold in recent years and the overall five-year survival rates remain dismal at 11% [1, 2]. Diagnosis at advanced stages, unclear molecular profiles, and few viable targeted therapeutics are among the factors causing high mortality rates [3, 4]. Moreover, most patients are present with underlying cirrhosis or decompensated liver disease which become difficult to treat with standard doses of chemotherapeutics [5, 6]. Drug resistance is another cause for therapy failure and is associated with the existence of tumor-like stem cells [7-9]. Yet mechanistic insight into pathways driving stem cell transformation, which could lead to targeted therapeutics, is limited for these cancers. Identification and validation of rational targets and signaling pathways underlying HCC development and progression are needed to develop better strategies for HCC diagnosis, prevention, and therapy.
Co-existing cirrhosis and drug resistance from a heterogeneous cancer are some causes for therapy failure: the latter is potentially associated with the existence of cancer stem cells. Therefore, identification and validation of rational targets underlying HCC development and progression- specific populations of HCCs such as those arising from cancer stem cells remain urgently needed for improved survival and decreased toxicities. Recently, we have observed that the TGF-β pathway is altered in 39% of HCCs and that the alterations correlate with raised high mobility group AT-hook 2 (HMGA2) and telomerase reverse transcriptase (TERT) levels [10]. TGF-β pathway members are downregulated in a significant number of human HCCs with a cancer stem cell signature [11, 12]. Additionally, recent TCGA meta-analysis has revealed a significant association between HMGA2 overexpression and poor overall survival in 14 types of cancers, including hepatocellular carcinoma [13]. HMGA2 is a small, non-histone, chromatin associated protein that lacks intrinsic transcriptional activity but regulates gene transcription by altering local chromatin structure at the promoter and/or enhancers [14, 15]. Increased evidence has suggested that HMGA2 could be involved in tumor growth [16-18], cancer cell differentiation [19, 20], and stem cell self-renewal [21, 22].
RING-finger E3 ligases are instrumental in the regulation of inflammatory cascades, apoptosis, and cancer. An increased abundance of negative regulators of the SMAD3-dependent tumor suppression, such as E3 ubiquitin ligases, represent a mechanism for the oncogenic function of the TGF-β pathway. PJA1 belongs to the PRAJA family of E3 ubiquitin ligases, whose expression has been shown to be elevated in some cancers including those of the gastrointestinal tract [23]. Overexpression of PJA1 in HCC cell lines promotes the ubiquitination of SMAD3 and β2SP [23]. PJA1 and SMAD3 respectively bind to SMAD3 and HMGA2. HMGA2 is induced by the TGF-β/SMAD pathway during EMT [24, 25]. To explore targets in HCC in the context of the TGF-β pathway, we analyzed genetic alterations of PJA1 in a significant number of HCCs and determined an interaction between PJA1 and HMGA2.
RESULTS
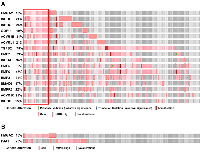
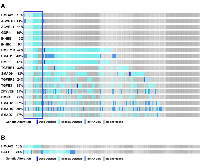
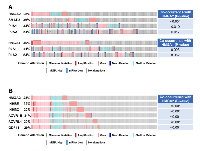
Our recent study has demonstrated that HMGA2 was overexpressed in samples with either mutations or amplifications in the TGF-β pathway genes [26]. In addition, the impact on the survival of cancer patients was most significant for overexpression of HMGA2, a collagen-encoding gene and MMP9. Therefore, we compared genetic alterations of HMGA2 and TGF-β pathway core genes in 348 HCC patients from TCGA database. Amplification, mutations, gains, and high mRNA of HMGA2 were compared with those of 43 TGF-β pathway core genes selected in our previous study [26]. Genetic alterations of 15 genes, INHBE, INHBC, GDF11, ACVR1B, ACVRL1, TGFB2, BMP7, INHBA, BMP5, BMP6, BMP2, SMAD5, BMPR2, ACVR1C, and INHBB, of the 43 TGF-β pathway core genes highly-moderately matched that of HMGA2 as highlighted by the red line box (Figure 1A). Although the gain and high mRNA of HMGA2 were compared with those of PJA1 in 348 HCC patients from TCGA analysis data, the genetic alterations lowly matched between the two genes (Figure 1B). Like Figure 1A, deletion and low mRNA of HMGA2 were compared with those of 43 TGF-β pathway core genes in 372 HCC patients from TCGA database. Genetic alterations of 17 genes, ACVR1B, ACVRL1, GDF11, INHBE, INHBC, BMPR1B, SMAD1, BMP3, TGFBR1, SMAD9, TGFBR3, TGFB3, ZFYVE9, BMP4, SMAD2, SMAD4, and SMAD7, of 43 TGF-β pathway core genes were shown to highly-moderately match that of HMGA2 as highlighted by the blue line box (Figure 2A). Deletions and low mRNA in generic alterations of HMGA2 moderately matched that of PJA1 in 372 HCC patients from TCGA database (Figure 2B). Co-occurrences of genetic alterations of PJA1/2 and TGF-β pathway core genes with that of HMGA2 were determined. Co-occurrences of mutation amplification, gain, deletions and high/low mRNA of HMGA2 with those of SMAD3, PJA1, and PJA2 in 348 HCC patients were significant with <0.001, 0.019, and 0.017 of P-values, respectively, while HMGA1 indicated high and low significant co-occurrences with PJA1 and PJA2, respectively (Figure 3A). Co-occurrences of mutation amplification, gain, deletions, and high/low mRNA of HMGA2 with those of 43 TGF-β pathway core genes in 348 HCC patients were highly significant in INHBE, INHBC, ACVR1B, ACVRL and GDF11 with all <0.001 of P-values (Figure 3B). These data indicate that the genetic alterations of HMGA2 concomitantly occurs with those of many TGF-β pathway core genes including PJA1/2 in HCC patients, suggesting that these concomitant genetic alterations of HMGA2 and TGF-β pathway core genes might be involved in tumorigenesis and development of HCC and thus, helpful as biomarkers for screening of HCC.
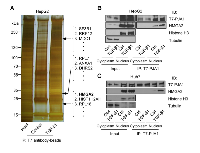
Next, we explored proteins that interact with PJA1 in HCC cells through a TGF-β dependent manner. Co-immunoprecipitations with T7-beads and SDS-PAGE of HepG2 cell lysate with T7-PJA1 overexpression with or without TGF-β treatment were performed. After silver staining of the gel, we found three protein bands with molecular sizes of approximately 200, 30, and 20 kDa specific in the cell lysate of HepG2 treated with TGF-β (Figure 4A). Each band was found, by mass spectrometry analysis, to include several proteins. Among the candidates of PJA1 interacting proteins, HMGA2 can bind to SMAD3, and PJA1 can also bind to SMAD3. In addition, HMGA2 is induced by the TGF-β/SMAD pathway during EMT [24, 25]. Taken togethe, we have considered that PJA1 could interact with HMGA2.
To confirm the interaction between PJA1 and HMGA2, co-immunoprecipitation and Western blotting were performed using cell lysates after cell compartment fractionation in HepG2 and Huh7. The data showed that PJA1 interacted with HMGA2 in the nuclear fraction of both HepG2 and Huh7 cells and that treating the TGF-β for 3 hours enhanced the interaction between HepG2 and Huh7 cells (Figure 4B).
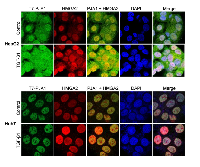
Next, confocal microscopy analysis was performed to investigate the co-localization of PJA1 and HMGA2 in HepG2 and Huh7 cells. The analyses demonstrated co-localization of PJA1 and HMGA2 in the nuclear of HepG2 and Huh7, and the co-localization was enhanced by TGF-β treatment for 3 hours (Figure 5).
DISCUSSION
HMGA2 protein is overexpressed in many types of cancer including oral squamous cell carcinoma [27], lung cancer [28], breast cancer [29], gastric cancer [30], liver cancer [31], pancreatic cancer [32], colorectal cancer [33], ovarian cancer [34], and endometrial cancer [35]. Human telomerase reverse transcriptase (hTERT) is found to be essential for tumor cell proliferation and self-renewal properties [36, 37]. HMGA2 modulates hTERT transcription to promote tumorigenesis [38]. HMGA2 interferes with the recruitment of HDAC2 to the hTERT proximal promoter through interaction with transcription factor Sp1, leading to localized histone H3K9 hyperacetylation and thereby stimulating hTERT expression and telomerase activity. Interestingly, HMGA2 knocked-down by sh-HMGA2 in HepG2 cells leads to the progression of telomere shortening and a concurrent decrease of steady-state hTERT mRNA levels. As supported by this view, our recent TCGA analyses have demonstrated that both HMGA2 and TERT expression levels were raised in the alterations of the TGF-β pathway genes in 33 cancer types [10]. Although further study is necessary, the regulation of TERT by HMGA2 might play a role in cancers that have disrupted TGF-β pathways, including HCC.
HMGA2 is a non-histone chromatin-binding protein which contains three AT-hook domains that enable its binding to the minor groove of AT-rich DNA stretches [39, 40] and allows organized protein complexes on enhancers of various genes to regulate gene expression and cell differentiation. Although the function of the C-terminal acidic region is poorly understood, there are implications that C-terminal acidic tail in HMGA1 is involved in both the protein-protein interaction and the recruitment of factors during regulation of gene transcription [41]. We found that HMGA2 can interact with PJA1 in HCC cells, with the interaction increased due to TGF-β treatment. HMGA2 seems to undergo ubiquitination through PJA1 and/or some other E3 ligase; however, TGF-β does not potentiate the ubiquitination (data not shown). Further study is needed to determine whether the HMGA2 interacts with PJA1 through the C-terminal tail, and to determine if the HMGA2 /PJA1 complex that binds to DNA and HMGA2-target genes are regulated by the interaction with PJA1 in TGF-β signaling in HCC cells.
HMGA2 was identified as a prominent TGF-β target in TGF-β–induced EMT of murine mammary epithelial NMuMG cells by transcriptomic analysis and has been shown to be induced by the SMAD3 pathway during EMT [24, 42]. TGF-β induces the expression of HMGA2 by activating the transcription factor SMAD. HMGA2 associates with SMAD complexes to induce Snail and Twist expression, two established regulators of EMT, leading to mesenchymal transition [43, 44]. Interestingly, the overexpression of HMGA2 enhanced TGF-β signaling by activating expression of the TGF-β type II receptor that is localized to the invasive front of tumors [25]. In addition, the studies have shown that Smad3 and PJA1 respectively bind to HMGA2 and Smad3. These data led us to consider that PJA1 might interact with HMGA2. The TGF-β signaling pathway has been strongly implicated in EMT induction for many cell types [24, 25, 42, 45]. Our data clearly shows that PJA1 interacts with HMGA2 in the nucleus of HCC cells, suggesting that PJA1 might translationally control EMT of HCC in the TGF-β signaling pathway through the interaction with HMGA2.
Since it was also demonstrated that HMGA2 regulates the TGF-β signaling pathway, future research should be carried out to elucidate how the interaction between PKA1 and HMGA2 participates in TGF-β signaling pathways of HCC cells. The function of HMGA2 as an oncoprotein may be associated with several important molecules involved in EMT, invasion, and metastasis of HCC cells. These results further indicate that HMGA2 may serve as a potential target for the development of therapies for HCC, although additional detailed studies in vivo are required.
MATERIALS AND METHODS
Cell culture and transfection
HepG2 (HB8065) from ATCC and Huh7 (gift from Dr. Aiwu Ruth He’s lab, Georgetown University) were cultured in DMEM/F-12 medium and supplemented with 10% fetal bovine serum. HepG2 and Huh7 cells were transfected with T7-PJA1 plasmid using Lipofectamine LTX (Invitrogen) according to the manufacturer’s instruction. TGF-β1 (Sigma, T1654) was added to create a final concentration of 200 pM. Human PJA1 was purchased from GeneScript (OHu55728D) and was subcloned into pcDNA3.1 T7 plasmid.
Mass-spectrometry analysis
HepG2 cells were transfected with T7-PJA1 plasmid and treated with or without TGF-β1 for three hours. The cell lysates were prepared with NP-40 buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1% NP-40, 1 mM EDTA) with proteinase inhibitor cocktail (Roche Applied Science) and 1 mg of the proteins were immunoprecipitated with T7 antibody-beads. After washing with NP-40 buffer, the samples were denatured with 2x Laemmli sample buffer by heating and were loaded on 4-15% gradient SDS-PAGE gel and silver-stained (Pierce, Silver Stain for Mass Spectrometry, 24600). Bands that were observed in TGF-β1 treated lane, but not in the control lane, were dissected from the stained gel and sent to Harvard Medical School for mass-spectrometry analysis.
Immunoblotting and immunoprecipitation analyses
Cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1% NP-40, 1 mM EDTA), protease inhibitor cocktail (Roche Applied Science), 1 mM PMSF, 1 mM NaF, and 1 mM sodium orthovanadate. Nuclear and cytoplasmic proteins were prepared as follows: cells were harvested and incubated in buffer A (10 mM Hepes, pH 7.8, 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 2 mg/ml aprotinin, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5% Triton X-100). After centrifugation, supernatants were collected as the cytoplasmic proteins. Buffer C (50 mM HEPES, pH 7.8, 420 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 2 mg/ml aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride) was added to the pellet. After rotation for 30 minutes and centrifugation, supernatants were collected as nuclear proteins. The following antibodies were used for immunoblotting and immunoprecipitation analyses: Flag-M2 (Sigma, F3165), Tubulin (T8328, Sigma), Histone H3 (sc-10809, Santa Cruz), T7 (A190-117A, Bethyl), HMGA2 (20795-I-AP, Proteintech), T7 Tag antibody agarose (69026, Novagen).
Confocal microscopy analysis
For confocal imaging, cells were plated onto coverslips in 6-well plates. After TGF-β treatment, the cells were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and blocked in 10% normal goat serum and PBS. The cells were incubated with primary antibodies, washed 3 times in PBS, and then incubated with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with Alexa-488 or Alexa-555 (Molecular Probes). 4’, 6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining. The slides were then examined using a Zeiss LSM 710 or Zeiss spinning disk confocal microscope and the images were acquired with the Zen 2009 software.
ABBREVIATIONS
HMGA2: High mobility group AT-hook 2; PJA: Praja ring finger E3 ubiquitin ligase; HCC: Hepatocellular carcinoma; TGF-β: Transforming growth factor-β; TCGA: The Cancer Genome Atlas; PBS: Phosphate buffered saline.
Author Contributions
LM - supervised the study designed, coordinated the research and intellectual input, KO experiments design, analyzed the data, and wrote the manuscript, JC-provided intellectual input, JS-data analytics.
ACKNOWLEDGMENTS
We acknowledge Emily kim for the careful reading.
CONFLICTS OF INTEREST
None.
FUNDING
This work was supported by the NIH grants: R01 AA023146 (L. Mishra), NIH R01 CA236591 (L. Mishra), NIH U01 CA230690 (L. Mishra), VA Merit I01BX003732 (L. Mishra) and GW CTR (L. Mishra).
- 1. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011; 365:1118–27. https://doi.org/10.1056/NEJMra1001683. [PubMed].
- 2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–86. https://doi.org/10.1002/ijc.29210. [PubMed].
- 3. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018; 15:599–616. https://doi.org/10.1038/s41571-018-0073-4. [PubMed].
- 4. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018; 68:723–50. https://doi.org/10.1002/hep.29913. [PubMed].
- 5. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016; 2:16018. https://doi.org/10.1038/nrdp.2016.18. [PubMed].
- 6. Dhanasekaran R, Bandoh S, Roberts LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000 Res. 2016; 5:F1000 Faculty Rev-879. https://doi.org/10.12688/f1000research.6946.1. [PubMed].
- 7. Thenappan A, Li Y, Kitisin K, Rashid A, Shetty K, Johnson L, Mishra L. Role of transforming growth factor beta signaling and expansion of progenitor cells in regenerating liver. Hepatology. 2010; 51:1373–82. https://doi.org/10.1002/hep.23449. [PubMed].
- 8. Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015; 524:180–85. https://doi.org/10.1038/nature14863. [PubMed].
- 9. Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RG, Grompe M, Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013; 494:247–50. https://doi.org/10.1038/nature11826. [PubMed].
- 10. Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, Kong AN. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila). 2008; 1:187–91. https://doi.org/10.1158/1940-6207.CAPR-08-0028. [PubMed].
- 11. Chen J, Zaidi S, Rao S, Chen JS, Phan L, Farci P, Su X, Shetty K, White J, Zamboni F, Wu X, Rashid A, Pattabiraman N, et al. Analysis of Genomes and Transcriptomes of Hepatocellular Carcinomas Identifies Mutations and Gene Expression Changes in the Transforming Growth Factor-β Pathway. Gastroenterology. 2018; 154:195–210. https://doi.org/10.1053/j.gastro.2017.09.007. [PubMed].
- 12. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, et al, and Cancer Genome Atlas Research Network. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018; 173:321–337.e10. https://doi.org/10.1016/j.cell.2018.03.035. [PubMed].
- 13. Huang B, Yang J, Cheng Q, Xu P, Wang J, Zhang Z, Fan W, Wang P, Yu M. Prognostic Value of HMGA2 in Human Cancers: A Meta-Analysis Based on Literatures and TCGA Datasets. Front Physiol. 2018; 9:776. https://doi.org/10.3389/fphys.2018.00776. [PubMed].
- 14. Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A, Manfioletti G, Giancotti V. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 2004; 574:1–8. https://doi.org/10.1016/j.febslet.2004.08.013. [PubMed].
- 15. Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007; 7:899–910. https://doi.org/10.1038/nrc2271. [PubMed].
- 16. Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, Wei JJ. Antiproliferative effects by Let-7 repression of high- mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008; 6:663–73. https://doi.org/10.1158/1541-7786.MCR-07-0370. [PubMed].
- 17. Hristov AC, Cope L, Reyes MD, Singh M, Iacobuzio- Donahue C, Maitra A, Resar LM. HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod Pathol. 2009; 22:43–49. https://doi.org/10.1038/modpathol.2008.140. [PubMed].
- 18. Cui H, Song R, Wu J, Wang W, Chen X, Yin J. MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am J Cancer Res. 2018; 8:405–21. [PubMed].
- 19. Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007; 104:11400–05. https://doi.org/10.1073/pnas.0704372104. [PubMed].
- 20. Tan L, Xu H, Chen G, Wei X, Yu B, Ye J, Xu L, Tan H. Silencing of HMGA2 reverses retardance of cell differentiation in human myeloid leukaemia. Br J Cancer. 2018; 118:405–15. https://doi.org/10.1038/bjc.2017.403. [PubMed].
- 21. Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008; 135:227–39. https://doi.org/10.1016/j.cell.2008.09.017. [PubMed].
- 22. Tzatsos A, Bardeesy N. Ink4a/Arf regulation by let-7b and Hmga2: a genetic pathway governing stem cell aging. Cell Stem Cell. 2008; 3:469–70. https://doi.org/10.1016/j.stem.2008.10.008. [PubMed].
- 23. Saha T, Vardhini D, Tang Y, Katuri V, Jogunoori W, Volpe EA, Haines D, Sidawy A, Zhou X, Gallicano I, Schlegel R, Mishra B, Mishra L. RING finger-dependent ubiquitination by PRAJA is dependent on TGF-beta and potentially defines the functional status of the tumor suppressor ELF. Oncogene. 2006; 25:693–705. https://doi.org/10.1038/sj.onc.1209123. [PubMed].
- 24. Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006; 174:175–83. https://doi.org/10.1083/jcb.200512110. [PubMed].
- 25. Morishita A, Zaidi MR, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y, D’Armiento J, Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013; 73:4289– 99. https://doi.org/10.1158/0008-5472.CAN-12-3848. [PubMed].
- 26. Korkut A, Zaidi S, Kanchi RS, Rao S, Gough NR, Schultz A, Li X, Lorenzi PL, Berger AC, Robertson G, Kwong LN, Datto M, Roszik J, et al, and Cancer Genome Atlas Research Network. A Pan-Cancer Analysis Reveals High- Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily. Cell Syst. 2018; 7:422–437.e7. https://doi.org/10.1016/j.cels.2018.08.010. [PubMed].
- 27. Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004; 64:2024–29. https://doi.org/10.1158/0008-5472.can-03-1855. [PubMed].
- 28. Sgarra R, Zammitti S, Lo Sardo A, Maurizio E, Arnoldo L, Pegoraro S, Giancotti V, Manfioletti G. HMGA molecular network: from transcriptional regulation to chromatin remodeling. Biochim Biophys Acta. 2010; 1799:37–47. https://doi.org/10.1016/j.bbagrm.2009.08.009. [PubMed].
- 29. Wu J, Zhang S, Shan J, Hu Z, Liu X, Chen L, Ren X, Yao L, Sheng H, Li L, Ann D, Yen Y, Wang J, Wang X. Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett. 2016; 376:284–92. https://doi.org/10.1016/j.canlet.2016.04.005. [PubMed].
- 30. Sun J, Sun B, Sun R, Zhu D, Zhao X, Zhang Y, Dong X, Che N, Li J, Liu F, Zhao N, Wang Y, Zhang D. HMGA2 promotes vasculogenic mimicry and tumor aggressiveness by upregulating Twist1 in gastric carcinoma. Sci Rep. 2017; 7:2229. https://doi.org/10.1038/s41598-017-02494-6. [PubMed].
- 31. Huang W, Li J, Guo X, Zhao Y, Yuan X. miR-663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother. 2016; 81:431–38. https://doi.org/10.1016/j.biopha.2016.04.034. [PubMed].
- 32. Rajamani D, Bhasin MK. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016; 8:38. https://doi.org/10.1186/s13073-016-0282-3. [PubMed].
- 33. Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, Hu S, Yao L, Peng J, Loera S, Xue L, Zhou B, Zhou L, et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res. 2011; 17:2570–80. https://doi.org/10.1158/1078-0432.CCR-10-2542. [PubMed].
- 34. Wu J, Liu Z, Shao C, Gong Y, Hernando E, Lee P, Narita M, Muller W, Liu J, Wei JJ. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011; 71:349–59. https://doi.org/10.1158/0008-5472.CAN-10-2550. [PubMed].
- 35. Wei L, Liu X, Zhang W, Wei Y, Li Y, Zhang Q, Dong R, Kwon JS, Liu Z, Zheng W, Kong B. Overexpression and oncogenic function of HMGA2 in endometrial serous carcinogenesis. Am J Cancer Res. 2016; 6:249–59. [PubMed].
- 36. Khattar E, Kumar P, Liu CY, Akıncılar SC, Raju A, Lakshmanan M, Maury JJ, Qiang Y, Li S, Tan EY, Hui KM, Shi M, Loh YH, Tergaonkar V. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J Clin Invest. 2016; 126:4045–60. https://doi.org/10.1172/JCI86042. [PubMed].
- 37. Beck S, Jin X, Sohn YW, Kim JK, Kim SH, Yin J, Pian X, Kim SC, Nam DH, Choi YJ, Kim H. Telomerase activity-independent function of TERT allows glioma cells to attain cancer stem cell characteristics by inducing EGFR expression. Mol Cells. 2011; 31:9–15. https://doi.org/10.1007/s10059-011-0008-8. [PubMed].
- 38. Li AY, Lin HH, Kuo CY, Shih HM, Wang CC, Yen Y, Ann DK. High-mobility group A2 protein modulates hTERT transcription to promote tumorigenesis. Mol Cell Biol. 2011; 31:2605–17. https://doi.org/10.1128/MCB.05447-11. [PubMed].
- 39. Winter N, Nimzyk R, Bösche C, Meyer A, Bullerdiek J. Chromatin immunoprecipitation to analyze DNA binding sites of HMGA2. PLoS One. 2011; 6:e18837. https://doi.org/10.1371/journal.pone.0018837. [PubMed].
- 40. Cui T, Leng F. Specific recognition of AT-rich DNA sequences by the mammalian high mobility group protein AT-hook 2: a SELEX study. Biochemistry. 2007; 46:13059–66. https://doi.org/10.1021/bi701269s. [PubMed].
- 41. Xu M, Sharma P, Pan S, Malik S, Roeder RG, Martinez E. Core promoter-selective function of HMGA1 and Mediator in Initiator-dependent transcription. Genes Dev. 2011; 25:2513–24. https://doi.org/10.1101/gad.177360.111. [PubMed].
- 42. Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial- mesenchymal cell transition. Mol Biol Cell. 2005; 16:1987–2002. https://doi.org/10.1091/mbc.e04-08-0658. [PubMed].
- 43. Tan EJ, Thuault S, Caja L, Carletti T, Heldin CH, Moustakas A. Regulation of transcription factor Twist expression by the DNA architectural protein high mobility group A2 during epithelial-to-mesenchymal transition. J Biol Chem. 2012; 287:7134–45. https://doi.org/10.1074/jbc.M111.291385. [PubMed].
- 44. Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008; 283:33437–46. https://doi.org/10.1074/jbc.M802016200. [PubMed].
- 45. Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994; 127:2021–36. https://doi.org/10.1083/jcb.127.6.2021. [PubMed].
Last Modified: 2020-06-09 18:30:56 EDT
