Genes & Cancer
Abstract | PDF | Full Text | Supplementary Materials 1 | Supplementary Materials 2
https://doi.org/10.18632/genesandcancer.207
Variant profiles of genes mapping to chromosome 16q loss in Wilms tumors reveals link to cilia-related genes and pathways
Eiko Kitamura1, John K. Cowell1, Chang-Sheng Chang1 and Lesleyann Hawthorn1
1 Georgia Cancer Center, Augusta University, Augusta, GA, USA
Correspondence to: Lesleyann Hawthorn, email: [email protected]
Keywords: Wilms tumor, chromosome 16q, cilia-related genes, cilia pathways
Received: July 06, 2020
Accepted: September 08, 2020
Published: October 06, 2020
Copyright: © 2020 Kitamura et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Background: Wilms tumor is the most common pediatric renal tumor and the fourth most common malignancy in children. Chromosome 16q deletion(del) or loss of heterozygosity (LOH) has been correlated with recurrence and overall poor prognosis, such that patients with 16qLOH and 1p allelic loss are treated with more aggressive chemotherapeutic regimens.
Methods: In the present study, we have compared the variant profiles of Wilms tumors with and without 16q del/LOH using both data available from the TARGET database (42 samples) and tumors procured from our legacy collection (8 samples). Exome-Seq data was analyzed for tumor specific variants mapping to 16q. Whole exome analysis was also performed. An unbiased approach for somatic variant analysis was used to detect tumor-specific, somatic variants.
Results: Of the 72 genes mapping to 16q, 42% were cilia-related genes and 28% of these were found to carry somatic variants specific to those tumors with 16qdel/LOH. Whole exome analyses further revealed that 30% of cilia-related genes across the genome carried alterations in tumors both with and without 16qdel/LOH. Additional pathway analyses revealed that many cilia-related pathway members also carried deleterious variant in these tumors including Sonic Hedgehog (SHh), Wnt, and Notch signaling pathways.
Conclusions: The data suggest that cilia-related genes and pathways are compromised in Wilms tumors. The genes on chromosome 16q that carry deleterious variants in cilia-related genes may account for the more aggressive nature of tumors with 16q del/LOH.
INTRODUCTION
Wilms tumor (WT) is the fourth most common pediatric cancer and affects approximately 1 in 10,000 children in Europe and North America. It typically presents as a complex embryonal tumor with triphasic histology (blastemic, epithelial and stromal components) and may also display cartilage, osteoid, and neural elements adding to the complexity of these tumors[1]. Although having a relatively good overall survival (>90%), due to a combination of surgery and more recently radiation/chemotherapy, there is also a subgroup of patients with poorer overall survival[2]. The stage at diagnosis is important to some extent in this determination, as is the histological subtype. WT show favorable (FHWT) or diffuse anaplastic (DAWT) histology, where the anaplastic histology is defined by the presence of atypical, polyploidy mitotic figures, large nuclei and hyperchromasia[3]. Bilateral tumors, usually associated with hereditary forms of the disease, cannot be treated with bilateral resection and therefore need alternative therapeutic strategies[4]. There is no apparent correlation between histology subtype and tumor stage, and only 50% of children that suffer relapse will survive. In addition, there is a high incidence of late radiation morbidity in patients undergoing adjuvant radiotherapy for Wilms tumor, significant adverse events and treatment-related risk factors in long-term Wilms tumor survivors and a high risk of second malignant neoplasms, presumed to be due to treatment.
Studies aimed at defining the molecular characteristics of relapsing WTs have identified abnormalities associated with poor outcomes including loss of heterozygosity (LOH) at 16q[5, 6].These observations have been confirmed by several groups in large studies in the National Wilms’ Tumor Study Group (NWTSG) study groups 3 and 4 as well as the United Kingdom Children’s Cancer Study Group (UKCCSG) studies[6, 7]. In the prospective NWTSG5 study, involving >2000 samples, 16% showed 16q LOH, and a significant correlation was found between poor prognosis and relapse within 2 years of treatment[5]. Additionally, LOH at 16q has been associated with a 2.7-fold increased risk of death in favorable histology tumors[6]. Moreover, a recent meta-analysis involving 10 studies and 3385 patients concluded that LOH at 16q was significantly associated with WT relapse in both histological subtypes[8]. These studies now clearly demonstrate that there is a subset of WT that are dependent on genetic events on 16q which determine poor outcome. Chromosome 16q and/or 1p allelic loss status are used to classify patients within the NWTSG therapeutic protocol to receive more rigorous chemotherapies[3, 5]. Knudson’s landmark ‘two hit hypothesis’[9] provided proof-of-principle that tumor suppressor genes could be identified by examining genes mapping to single allele deletions accompanied by variants in second alleles at these loci. However, attempts to identify loci at chromosome 16q have largely been futile due to the extensive regions of LOH typically observed, possibly suggesting that several genes may act in concert to contribute to the more aggressive phenotype.
We have used data generated through the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative, which has enabled comprehensive characterization of high-risk Wilms tumor cases, defined as having either favorable histology (FHWT) that relapsed or diffuse anaplasia (DAWT). These samples were combined with a legacy set of samples to define variant-containing genes on chromosome 16 which may contribute to Wilms’ tumorigenesis or the more aggressive nature of tumors with 16q del/LOH. As a result, it became apparent that a large percentage of the tumor-specific variants detected in WT with 16q loss and/or LOH affected genes that encoded cilia- related proteins. A more extensive genome analysis beyond chromosome 16 demonstrated tumor-specific, variants in additional genes associated with ciliogenesis.
RESULTS
Microarray analysis of copy number and LOH
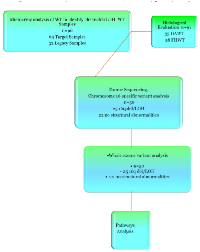
Figure 1 shows the experimental layout in order to clarify the number and types of samles used in each of the experiments. In our previous study[10] we defined three types of 16q abnormality; deletion with accompanying LOH, LOH without 16q deletion and copy number abnormalities (CNA) without LOH, presumably resulting from chromosome loss from a tetraploid cell. Of 69 paired samples from the TARGET project, 32 (46%) carried either deletions or LOH involving 16q, which included 24 samples with deletion associated with a LOH (35%) and 8 samples (11%) carrying deletions without accompanying LOH (Figure 2).
Deletions with accompanying LOH extended the entire length of 16q, while deletions without accompanying LOH, localized to chr16q11.2 (46506039-46539392) and 16q24.3 (90158838-90287536). These tumors are now collectively termed 16qdel/LOH samples. Combining the TARGET data with that previously reported (Hawthorn and Cowell 2011), 96 samples were available for analysis (Supplemental Table 1).
Within this series of tumors, both histological evaluation and assessment of chromosome 16q status was available from 91 cases, consisting of 33 with DAWT and 58 with FHWT. 60% (20/33) of tumors with DAWT histology and 36% (21/58) with the FHWT histological subtype had loss/deletion of chromosome 16q . Figure 3 shows that structural abnormalities of 16q are significantly associated with the DAWT histology (Welch 2-sided t-test p = 0.02).
Variants detected in WT
Exome-Seq data from 50 WT samples was analyzed, 42 samples from the TARGET project and 8 samples from the legacy collection. The annotated variants were filtered for predicted functional effects as outlined in the Materials and Methods section and categorized into missense variants, frameshifts, deletions/insertions and stop gain/losses. A total of 24,271 heterozygous-/homozygous- somatic variants predicted as Damaging and/or Deleterious were identified in 10,789 genes. The full list of somatic variants is available in Supplemental Table S2. It should be noted that the number of variants are higher than other published works due to the fact that we did not filter out variants that were listed in dbSNP but instead focused on variants that were present in the tumors and not in the matched normal samples, ie “tumor-specific/somatic”.
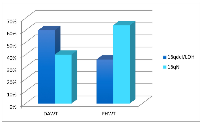
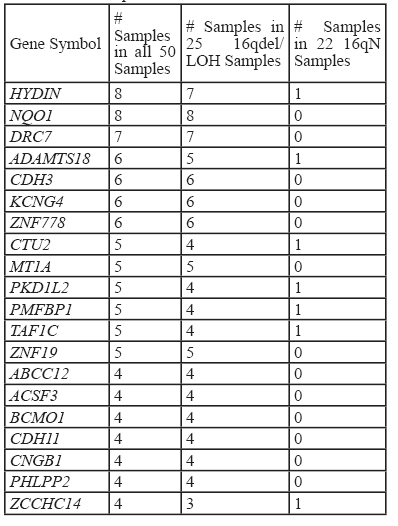
To examine the variant profiles in the 16qdel/LOH samples, exome data from 50 samples were analyzed, 25 of which were16qdel/LOH and 22 with no structural abnormalities of 16q and three that had no accompanying copy number data available. Using the pipeline described in the Materials and Methods section, we identified 131 homozygous or hemizygous in the samples with allelic loss, somatic variants in 91 genes that mapped to 16q in the 16qdel/LOH samples. Chromosome 16N tumors carried somatic variants in 13 genes, 10 of which overlapped with the variants in the 16qdel/LOH samples and 3 genes that were unique. The 16qdel/LOH group carried a median of 8.1 (homozygous or hemizygous in the samples with allelic loss, deleterious/damaging) variant-containing genes mapping to chromosome 16q per patient, while 16qN group carried a median of 0.59 genes per patient (See Figure 4). The top 20 variant-containing genes are shown in Table 1. The complete list of all somatic variants mapping to 16q in tumors with and without 16q abnormalities is available in Supplemental Table S3.
As envisaged, the number of variants-containing genes mapping to chromosome 16q is higher in the DAWT group. Figure 4b shows that there is also a statistically significant difference (p = 0.07) between the number of variant-containing genes carried by the patients with DAWT (median= 5.8 genes/patient) compared with patients with FHWT (median=2.7 genes/patient). There are 33 genes overlapping between the 2 histological subtypes, 31 uniquely altered in the DAWT and 18 unique to the FHWT subtypes.
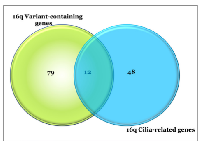
Analysis of gene classification using GO Genesets in GSEA revealed that GO-Cilium was significantly (q = 3.5e-2) enriched in the 91 variant-containing genes mapping to 16q. The CilDB was then queried using chromosome location and 60 genes were identified that mapped to chr16q. The 91chromosome 16q variant-containing genes were then cross-referenced with those in the CilDB which identified 12 that overlapped (see Figure 5). Of note, however, many of these genes are included in the CilDB as a result of genome-wide transcript expression analyses and have not been fully validated as having cilia-related functionality. We, therefore, further curated genes that had also been reported in the literature as ‘cilia-related’, by GO functional categorization and the DAVID functional annotation tool.
Table 2 shows 12 of the cilia-related genes which contain somatic variants. Notably, 17/25 16qdel/LOH samples carry variants in cilia-related genes mapping to 16q. It is noteworthy that only one patient without a 16q abnormality carried homozygous somatic variants in any of these genes.
Whole exome analysis of WT
Given the overwhelming number of cilia-related genes on chromosome 16q that carried somatic variants, we extended the variant profiles of cilia-related genes to the entire exome and also determined whether there was a difference in variant profiles between tumors with (n = 25) and without (n = 22) 16q structural abnormalities. 23,429 variants were detected in the 47 tumor samples that were predicted to be detrimental at the protein level as outlined in the Materials and Methods. A total of 4960 and 4679 genes respectively carried somatic variants in two or more samples with and without 16q abnormalities and 3577 genes overlapped between the two groups. Of these, 1383 and 1102 genes respectively were unique to the tumors with and without 16q abnormalities. Table 3 shows the 30 most frequently altered genes for each of these groups. The complete list of somatic variants is available in Supplemental Table 2.
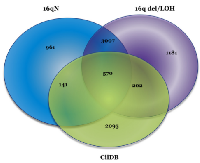
Analysis using the CilDB showed a large number of genes overlapping with cilia-related functions from tumors with and without 16q abnormalities and these findings are shown in Figure 6, where it can be seen that 570 genes in both sample types overlap with CilDB genes, 141 are unique samples with no 16q abnormalities and 202 uniquely overlap between the 16qdel/LOH and the CilDB. The genes that comprise each of the overlaps are defined in Supplemental Table S4A.
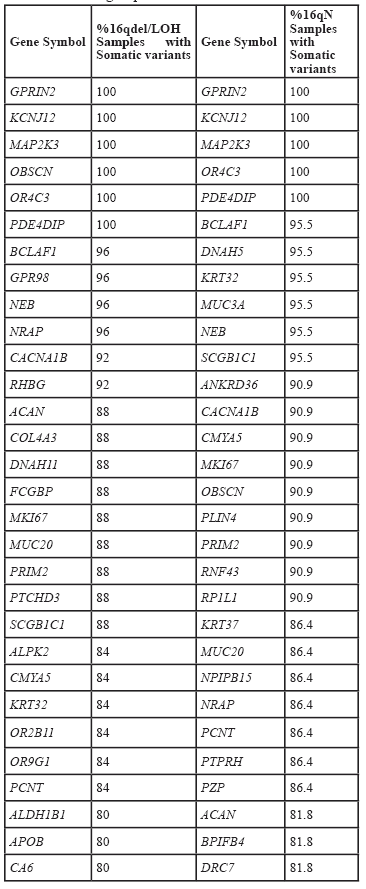
Interestingly, genes that carried variants in the 16qdel/LOH samples were identified by this analysis as having deleterious variants in tumors with and without 16q abnormalities. For example, the HYDIN gene had homozygous, somatic variants in 7/25 16qdel/LOH samples but only 1/22 homozygous variants were detected in the 16qN samples (Table 2). Using whole exome analysis, however, 12/25 and 11/22 somatic variants were detected in the two groups respectively, however not all of these were homozygous (or hemizygous in the samples with allelic loss) variants (Supplemental Table 4B).
The 570 variant-containing genes identified in the CilDB and overlapping between tumors with and without 16q abnormalities (Figure 6) were also analyzed using DAVID and GSEA to refine the list of cilia- related genes. Using the functional annotation clustering algorithm in DAVID the most highly enriched annotation cluster included cilia biogenesis, cilium assembly and cilium morphogenesis with an enrichment score of 14.04. Other annotation clusters included with high enrichment scores included cilium movement/primary ciliary dyskinesia (ER=7.7), dynein heavy chain/ axonemal dynein complex (ER=7.47) and intraciliary transport/primary cilium (ER=3.1). Using GSEA on the same 570 overlapping genes (with an FDR q value of <0.05), 266 genes were identified as belonging to the GO classifications of GO_ CILIUM , GO_ CILIUM MORHOGENESIS, GO_ CILIUM ORGANIZATION, GO_ CILIARY PART, GO_ CILIARY PLASMA, GO_ PRIMARY CILIUM, GO_ MEMBRANE, GO_ AXONEME, GO_ AXONEME ASSEMBLY, GO_ CENTROSOME, GO_ CENTRIOLE. One hundred and forty-three of the 266 carried variants in at least 25% of 16qdel/LOH samples and 149 in 25% of the tumors without 16q abnormalities.
Established cilia-dependent pathways
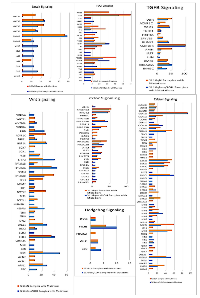
We next examined well-established, cilia-dependent pathways including Sonic Hedgehog (SHh), Wnt, Platelet-derived growth factor receptor (PDGFR), Notch, TGF-B and mTOR for variants in their component molecules. The pathway members and the percentage of samples with variants in tumors with and without 16q abnormalities are shown in Figure 7.
Although there is a large degree of overlap in the variants for each pathway, some of the genes in each of these pathways differ. For example, in the Wnt Signaling pathway APC, BCL9, KREMEN, c-JUN, FDZ9 and MDM2 variants are evident in the 16qdel/LOH samples exclusively and conversely, AXIN, CDKN2A, SOX7 and WNT11 are altered solely in the tumors without 16q abnormalities. The SHh pathway shows larger percentages of somatic variants in the PTCH1 and GLI3 genes in the 16qdel/LOH samples, while PIK3R1, ATM and PLD genes are more frequently altered in tumors without 16q abnormalities. All pathways are shown in Supplemental Image 1 (SI1).Overall, a down-regulation of these pathways due to inactivating somatic variants is predicted.
Variant containing genes previously reported in Wilms tumor
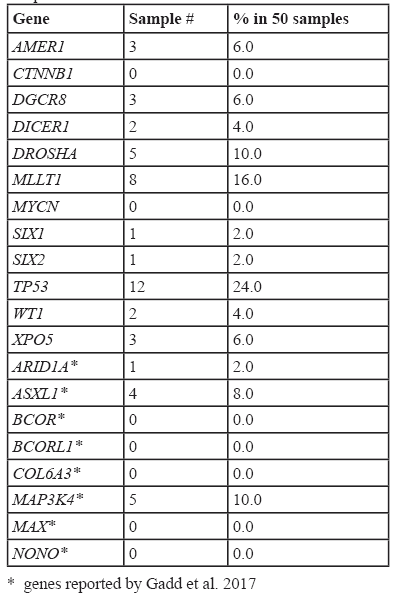
Genes that have been previously reported to carry variants in Wilms tumors also were found to carry variants in this study, however using our analysis pipeline these were not determined to be frequent events. For example, in AMER1, we detected somatic variants in 6% of samples, CTNNB1 (0%), DICER1 (4%), DGCR8 (6%), DROSHA (10%), MLLT1 (16%), SIX2 (2%) and TP53 (24%). The list of the most frequently reported WT-associated variant-containing genes is shown in Table 4.
DISCUSSION
Loss of heterozygosity involving distinct regions of chromosome 16q been extensively correlated with poor outcome in Wilms’ tumors and is now included as a test in evaluating therapy options for WT patients. Although the incidence of 16q loss in WT tends to be 15-20%, [1, 10, 14]the present study a much higher frequency (49%) was observed, most likely due to the TARGET samples being preselected as high-risk, i.e. having either favorable histology (FHWT) that relapsed or DAWT. Wittman et al[15] also reported a higher incidence of 16q allele loss in mixed-type as well as in diffuse anaplastic tumors, whereas epithelial and stromal tumors rarely exhibited 16q losses. It was also notable that chromosome 16 loss/ LOH was a less frequent event in the FHWT class and the majority were classified as DAWT and the number of variant–containing genes mapping to chromosome 16q was higher in the DAWT as opposed to the FHWT histologically classified samples. It is interesting that Gadd et al[13] reported a higher percentage of overall variants across the genome in DAWT samples using the TARGET data.
The identification of genes mapping to chromosome 16q that play a role in WT tumorigenesis have been largely ineffectual and has led to the assumption that more than one gene mapping to this region could attribute to the more aggressive nature of tumors carrying 16q deletion or LOH. The observation of LOH is generally taken as being a mechanism of exposing recessive variants in gene critical to tumorigenesis and through our variant profiles focusing on 16q we found a remarkably consistent incidence of somatic variants in genes related to cilia structure and function. WT is considered an embryonal tumor arrested in early stages of kidney development. An early stage of this process involves the two-way induction of differentiation induced by the contact of the ureteric bud with the metanephric mesenchyme generating waves of differentiation signals as the bud invades the blastemal mass. During this process the kidney goes through differentiation into primitive stages of a pro-nephros and a meso-nephros which are normally degraded to give rise to the metanephric kidney[16]. Importantly, cilia have been shown to play an important role in early stages of this process[17] and ciliary structures can be seen in Wilms tumors reflecting their early stage in developmental arrest[18]. The frequent involvement of somatic variants in genes that are related to cilia, therefore, potentially represents a mechanism of sustaining embryonic status of cells in WT.
The large number of cilia-related genes both mapping to chr16q and containing variants in the 16qdel/LOH samples aroused our interest in the role of cilia in WT. Specifically we focused on 12 genes that were highly referenced in the literature. Out of 25 16qdel/LOH samples, only 8 did not have variants in cilia-related genes. Recent studies have shown that hundreds of proteins reside permanently or transiently in cilia. In the kidney, immotile or primary cilia are present on the apical (urinary /luminal) surface of epithelial cells from all tubular segments and are critical sensory and signaling centers. Primary cilia are solitary and immotile cellular appendages that serve as signaling hubs for many signaling pathways during development (see below). Defects in their structure/function result in a spectrum of clinical and developmental pathologies.
Some of the genes that were detected through our analysis are related to motile cilia components and would not impact the primary or non-motile renal cilium, however it should be kept in mind that these tumors arise in the developing kidney and are blastemal in nature. Furthermore, the role of motile cilia in nephrogenesis has not been elucidated, for example, multi-ciliated cells, or cells that contain multiple motile cilia have been reported in fetal kidney tubules[19] and sporadically in an assortment of renal diseases[20, 21]. Additionally, despite the distinction between primary and motile cilia, it has been reported that there is a clinical overlap between Primary Ciliary Dyskinesia (PCD) and many of the non-motile ciliopathies[22].
The most frequently altered gene detected in the present study was DRC7 (dyenin regulatory complex 7) a misnomer as this is a conserved ciliary protein localized to the outer microtubule doublets of the axoneme and is not associated with the dynein arms[23]. The role of this protein localizing to the axoneme implies that it may also be involved in primary cilia function, and play a role in nephrogenesis.
Several of the detected somatic variants mapping to 16q are involved in PCD a rare genetic disease that is inherited as an autosomal-recessive trait. For example, HYDIN was frequently found to carry variants in patients with chromosome 16q del/LOH. This gene encodes a protein that is integral to the central pair apparatus of motile cilia. Raidt et al[24]examined ciliary beat patterns for PCD patients carrying different somatic variants and reported that many of the cilia in nasal brushings of HYDIN mutation carriers were actually primary/immotile cilia, suggesting that the classification of these two subtypes of cilia is not definitive.
Another frequently altered gene was NQO1, a gene which has been reported as down-regulated in PCD biopsies of lung[25] and reduced expression leads to increased kidney injury in response to cisplatin[26].
GAS8, another PCD-related gene, encodes a subunit of the nexin-dynein regulatory complex and connects microtubule doublets. GAS8 variants are associated with axonemal disorganization sometimes characterized by a partial loss of inner dynein arms[27, 28]. Interestingly, although GAS8 localized to the microtubule axoneme of motile cilia it also localized to the base of non-motile/primary cilia[29] and the role in non-motile cilia has not been elucidated. Intriguingly, GAS8 plays a role in the SHh signaling pathway. Evron et al[30] have shown in a murine model that Gas8 binds to Smoothened (Smo) and acts at the base of primary cilia as a regulator of Smo entry into the cilium following SHh pathway activation. In the absence of Gas8, Smo accumulation in the cilium is abrogated and that it cannot activate the Gli transcription factors that ultimately govern the expression of downstream genes.
The transition zone (TZ) is the proximal-most domain of the ciliary axoneme, found immediately distal to the basal body and is critical to cilium formation and functions as a portal that maintains the correct composition of the ciliary organelle. Variants in genes that affect TZ function result in wide range of ciliopathies. TMEM231 is mandatory for the localization of a subset of the MKS complex components to the TZ and to maintain ciliary protein composition. Another TZ protein, RPGRIP1L is also mutated in Merkel Syndrome (MKS) which is characterized by kidney cysts. Furthermore, variants in this gene cause Nephronophthisis (NPHP) which also characterized by kidney cysts. Genetic disruption of the transition zone disorders the ciliary localization of membrane-associated proteins including SHh-related SMO that requires the transition-zone proteins including TMEM231 to accumulate within the ciliary membrane. Consequently, loss of any of these proteins leads to SHh-associated developmental defects[31].
Whole exome analysis also revealed that a large percentage of genes associated with ciliogenesis and mitosis carried somatic variants in both 16qN and 16qdel/LOH samples. Given that WT is a developmental tumor, these findings are notable. Although not reported specifically for Wilms tumor, an ever-increasing number of papers report on a decrease, loss, or distortion of the primary cilium in a variety of cancer types. It is commonly assumed that the cilium act as to control cellular proliferation by employing the same structural components required for chromosome segregation[32]. Loss of the cilium in cancer cells may, therefore, result in loss of these components and contribute to distorted cellular signaling[33]. Additionally, given the critical role of cilium in cell division, defects in cilia- related genes have become a viable explanation for ciliopathy phenotypes that appear during development as many cells are actively proliferating during this phase and therefore do not have cilia[34].
Further analysis of the established cilia-related pathways revealed that a large number of genes in these pathways carried somatic variants in the WT samples. The SHh was discussed above in the context of 16qdel/LOH homozygous variants detected in the GAS8 and TMEM231 genes. The SHh signaling plays an essential role in many aspects of embryonic development and tumorigenesis. The cilium functions as the transduction hub for SHh signaling[35]. Our analysis found 34% of samples carried PTCH1 somatic variants and somatic variants of GLI3, GLIS1 STK3 and PRKAG2 were also detected. Variants in PTCH1 are associated with rhabdomyosarcoma[36], other studies report increased expression of PTCH1 in pediatric solid tumors including Wilms tumor[37].
Many parallel and divergent lines of evidence point to Wnt Signaling (both canonical and non-canonical) as central pathways in nephrogenesis. The Wnt/β-catenin pathway is one of the multiple signaling pathways that cooperate in the initiation and progression of mesenchymal-epithelial transition and several members of the Wnt family have been implicated in the induction of epithelial renal vesicles (See 38 review). Wilms tumor development is tightly linked to nephron genesis and are frequently found within nephrogenic nests that resemble embryonic structures suggesting a block in the nephrogenic process by variants in the Wnt Signaling members. We report a large number of genes involved Wnt Signaling are altered in the WT cohort examined. The cilia and basal body function as regulatory mechanisms to govern Wnt signaling and signaling is mediated in the primary cilia. Additionally, ciliary-related proteins have been also shown to regulate Wnt signaling pathways.
Notch signaling also plays an elemental role in mammalian nephrogenesis. The differentiation of nephron progenitors requires the down-regulation of SIX2 and this accomplished by the Notch Signaling pathway[39]. Notch also regulates the process of nephron segmentation involving the differentiation of progenitor cells into the renal corpuscle, proximal tubule, loop of Henle and distal tubule[40]. Cells lacking the Notch signaling pathway fail to form these structures reminiscent of the disorganized nephrons typically observed in WT. The Notch signaling pathway plays a central role in left right symmetry and cilium length control. Our analysis revealed somatic variants in all 4 Notch receptors, with 23% of samples carrying somatic variants in NOTCH3.
The plethora of events, both in normal and disease states, involving calcium signaling is overwhelming, however studies of the role of calcium in kidney development has historically been largely overlooked. PKD1 and PKD2 encode proteins that interact to form a calcium permeable channel in response to mechano-sensory stimuli in ciliary membrane and this channel becomes dysfunctional due to variants in PKD patients. Other interacting members of PKD2, TRPC1 and TRPV4 show strong expression patterns in the embryonic proximal tubules and ureteric bud and genes carried somatic variants in our cohort. Additionally, calcium entry in response to extracellular stimuli results in calcineurin (PPP3CA) activation, and signal transduction from the cytoplasm into the nucleus through dephosphorylation and nuclear translocation of the transcription factor nuclear factor of activated T cells (NFAT). This initiates a cascade of transcriptional events involved in physiological and developmental processes[41]. We detected a large number of variants in members of the NFAT family with 55% of patients carrying somatic variants in NFAT1. Notably, CABIN1, a calcineurin binding protein which results in decreased PPPC3A expression is highly expressed in mesenchymal progenitor cells at the onset of metanephric kidney development has been found to be over-expressed in WT[42]. In the present study, we detected CABIN1 somatic variants in 28% of samples assayed.
Other established cilia-related pathways are less documented in terms of nephrogenesis or WT. Notwithstanding, the mTOR signaling pathway is mediated by the primary cilia and inappropriately activated in cyst-lining epithelial cells in human ADPKD patients and mouse models[43]. Defects in the autophagy pathway have been detected in PKD and ciliopathies display impaired autophagy[44]. TGFβ and PDGF signaling are intricately involved in the development of renal fibrosis[45].
Whole exome analysis concurred with similar studies using the target dataset with some major differences. The most frequently altered genes in our study did not correspond to those reported by Gadd et al[13]. This was primarily due to differences in data filtering for instance, their discovery set was limited to variants reported in COSMIC including nonsense, and frameshift variants and verified somatic missense and in-frame variants predicted to be damaging and not identified in 1000 Genomes series 3. We did not filter our data using these criteria but relied on detecting somatic variants that were tumor specific. As a result, our somatic variant list is much more extensive and we detected many genes that were altered in a large percentage of samples, however our sample size is smaller.
TP53 was the most commonly mutated gene in a study reported by GADD et al[13] using their discovery set analysis, the mutation rate was 22% and using our analysis, we calculated the variant-containing rate as 26% (Table 4). Similarly, we found our variant-containing rates concurred with those reported by Gadd et al for most of the WT-related genes except for CTNNB1, a gene reported by Gadd et al, as the second most frequently variant-containing gene whereas we detected no variants using our analysis pipeline. Presumably, the predictions for the effects of somatic variants by PolyPhen and SIFT were finally selected to be damaging and/or deleterious in our study.
MATERIALS AND METHODS
Copy number analysis
We have previously performed copy number analysis for the 8 legacy Wilms tumor samples reported by Cowell and Hawthorn[10]. The TARGET samples were analyzed using Partek Genomics Suite 6.6 (Partek Inc., St Louis, MO, USA) for LOH and copy number alterations (CNAs) using paired .CEL and .CHP files, which were generated using Genome-Wide Human 6.0 SNP arrays (Affymetrix), from the TARGET database. For CNA analysis, the genomic segmentation algorithm available in Partek Genomics Suite was employed using default parameters, including minimum genomics markers: 50, signal to noise ratio: 0.3, segmentation p-value: 0.01. LOH was analyzed using the Hidden Markov model (HMM) algorithm with default parameters, max probability: 0.9999 and genotype error: 0.01.
DNA samples for exome sequencing
Genomic DNAs were prepared from 8 WT tissues (GOS100, 11, 132, 32, 52, 54, 576, 900), using standard phenol/chloroform extraction procedures or Wizard Genomics DNA purification kits (Madison, WI). Tumor samples were collected immediately following surgical resection during the period 1982–1992 and snap frozen in liquid nitrogen. At the time of collection, the diagnosis of WT was confirmed histopathologically, although in some cases the specific stage was not recorded. Unfortunately, it has also not been possible to recover this information retrospectively from some of the anonymized legacy samples. DNA was prepared from the whole tumor sample from snap frozen tissue using standard phenol/chloroform extraction procedures. Supplemental Table S1 shows the clinical information available for each sample used in the study and specifies which analyses were performed on the individual samples.
Exome sequencing
Exon capture libraries were constructed from 1 ug of genomic DNA using the Agilent Human All Exon Target Enrichment kit (38 Mb) or the SureSelect Human All Exon 50 Mb kit according to the manufacturer’s protocol (Agilent, Santa Clara, CA). The individual libraries were quality-checked and quantified using the Agilent 2100 Bioanalyzer and SYBR Green-qPCR (BIO-RAD, Hercules, CA), respectively. The libraries were sequenced on the Illumina HiSeq 2500 ver.2 (Illumina, San Diego, CA) using paired-end, 50 bp cycles. Base calling, reads quality assessment, de-multiplexing, and transferring to a fastq format was performed using Illumina data analysis software. FastQC was used for Quality control of the sequence reads. The average number of reads was 50 million/sample and on average, 92 % of total reads uniquely mapped on the NCBI37/hg19 reference sequence.
Exome data analysis
Reads that passed quality control were then aligned to the human reference genome (hg19) using BWA (Burrow-Wheelers Aligner version 0.6.1) with default settings. The generated BAM files were imported into Genome Analysis Toolkit, GATK2, for removal of duplicates, local realignment, mate-pair fixation, re-calibration and then variants were detected by HaplotypeCaller with a cutoff of depth at least 5. Germline variants were excluded by subtracting normal genotypes from tumor profiles using bedtools. In the case of our 8 legacy samples, we had lymphblastoid cell lines from 3 patients, for the remaining 5 patients we used the pooled lymphoblastoid cell line data as a baseline. The somatic variants resulting from these analyses were annotated using Ensemble variant effect predictor, VEP (release ver. 75). In the same way, 42 matched tumor-normal WTs, available through the TARGET study, were analyzed to search for somatic variants on chr16q. These annotated data were then filtered to investigate only those variants that were predicted to cause protein dysfunction and included deletions and insertions that lead to frame shifts, variants in critical splice junction nucleotides and missense variants generating stop codons or leading to predicted deleterious events in the protein. SIFT (Sort Intolerant From Tolerant)[11] and/ or PolyPhen-2 (Polymorphism Phenotyping v.2.)[12]were used to annotate the damaging and deleterious effects of missense variants.
For analysis of 16q specific somatic alterations, the data were filtered to include only altered alleles on chr16q. The annotated variants from exome data were categorized according to the chr16q structural abnormalities as determined by microarray analysis resulting in 25 WTs with16qdel/LOH and 22 WTs with wild-type chr16q profiles (16qN). For 16qdel/LOH samples, variants caused by deletion were sorted according to the ratio of read depths between a reference and an altered allele for each position and filtered to include variants with >=80% of altered allele ratio in tumor samples and <= 75% in the matched control samples with consideration for normal tissue contamination.
Several pathway analysis programs were used to analyze the data, including Ingenuity Pathway Analysis (IPA, http://www.qiagenbioinformatics.com), Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8.https://david.ncifcrf.gov/home.jsp), Geneset Enrichment Analysis (GSEA, http://software.broadinstitute.org/gsea/msigdb/annotate.jsp). Analysis of cilia-related gene overlaps was conducted using CilDB (http://cildb.cgm.cnrs-gif.fr). For DAVID, a Fisher Exact P-Value is used to determine whether the proportion of genes falling into each category differs by group. The Fisher Exact is adopted to measure gene enrichment in annotation terms. We have reported Enrichment scores for each of the Functional Annotation clusters which is based on the p-values for each of the term members. An FDR q-value=0.05 was used to compute overlaps in GSEA Hallmark Genesets.
CONCLUSION
In summary, we have found that 49% of WT samples carry a del/LOH event on chromosome 16q when the patient cohort is preselected as high risk. These findings support a number of other studies implicating structural abnormalities of 16q with more aggressive form of WT. Studies aimed at identifying causative genes in minimal regions of overlap in patients with del/LOH at 16q have been largely unsuccessful and this may be due to multiple gene variants mapping to that region of the genome. We have identified a series of WT samples with chromosome16q del/LOH and defined deleterious variants in genes mapping to that region. We propose that increased ciliary dysfunction may be responsible of the more aggressive nature of WT with chromosome 16q del/LOH.
ABBREVIATIONS
| Acronym | Description |
|---|---|
| 16N | tumors with no loss of 16q |
| 16qdel/LOH | Tumors with deletion or LOH of 16q |
| ABCC12 | ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| ACAN | aggrecan |
| ACSF3 | acyl-CoA synthetase family member 3 |
| ADAMTS18 | DAM metallopeptidase with thrombospondin type 1 motif, 18 |
| ADPKD | Autosomal Dominant Polycystic Kidney Disease |
| ALDH1B1 | aldehyde dehydrogenase 1 family, member B1 |
| ALPK2 | alpha-kinase 2 |
| AMER1 | APC membrane recruitment protein 1 |
| ANKRD36 | ankyrin repeat domain 36 |
| APC | Adenomatous polyposis coli |
| APOB | apolipoprotein B |
| ARID1A | AT rich interactive domain 1A |
| ASXL1 | additional sex combs like 1 |
| ATM | ataxia telangiectasia mutated |
| AXIN | Axis Inhibition Protein |
| BCL-9 | B-Cell Lymphoma-9 |
| BCLAF1 | BCL2-associated transcription factor 1 |
| BCMO1 | beta-carotene 15,15’-monooxygenase 1 |
| BCOR | BCL6 Corepressor |
| BCORL1 | BCL6 Corepressor Like 1 |
| BPIFB4 | BPI fold containing family B, member 4 |
| BWA | Burrow-Wheelers Aligner |
| c-JUN | Jun Oncogene |
| CA6 | carbonic anhydrase VI |
| CABIN1 | calcineurin binding protein 1 |
| CACNA1B | calcium channel, voltage-dependent, N type, alpha 1B subunit |
| CCDC135 | coiled-coil domain containing 135 |
| CDH11 | cadherin 11 |
| CDH3 | cadherin 3 |
| CDKN2A | cyclin-dependent kinase inhibitor 2A |
| CES1 | carboxylesterase 1 |
| CHST5 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| CMYA5 | cardiomyopathy associated 5 |
| CNA | Copy Number Alteration |
| CNGB1 | cyclic nucleotide gated channel beta 1 |
| COL4A3 | collagen, type IV, alpha 3 |
| COL6A3 | Collagen Type VI Alpha 3 Chain |
| CTNNB1 | catenin (cadherin-associated protein), beta 1 |
| CTU2 | cytosolic thiouridylase subunit 2 homolog |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
| DAWT | Diffuse Anaplastic Wilms Tumor |
| del | deletion |
| DGCR8 | DGCR8 microprocessor complex subunit |
| DICER1 | dicer 1, ribonuclease type III |
| DNA | Deoxyribonucleic Acid |
| DNAH11 | dynein, axonemal, heavy chain 11 |
| DNAH5 | dynein, axonemal, heavy chain 5 |
| DRC7 | dyenin regulatory complex 7 |
| DROSHA | drosha, ribonuclease type III |
| ENKD1 | enkurin domain containing 1 |
| FCGBP | Fc fragment of IgG binding protein |
| FDZ9 | Fizzled 9 |
| FHWT | Favorable Histology Wilms Tumor |
| GAS8 | growth arrest-specific 8 |
| GATK | Genome Analysis Toolkit |
| GLI3 | GLI family zinc finger 3 |
| GLIS1 | GLIS family zinc finger 1 |
| GPR98 | G protein-coupled receptor 98 |
| GPRIN2 | G protein regulated inducer of neurite outgrowth 2 |
| GSEA | Geneset Enrichment Analysis |
| HMM | Hidden Markov Model |
| HSD17B2 | hydroxysteroid (17-beta) dehydrogenase 2 |
| HYDIN | axonemal central pair apparatus protein |
| IPA | Ingenuity Pathway Analysis |
| KCNG4 | potassium voltage-gated channel, subfamily G, member 4 |
| KCNJ12 | potassium inwardly-rectifying channel, subfamily J, member 12 |
| KREMEN | Kringle domain-containing transmembrane protein |
| KRT32 | keratin 32 |
| KRT32 | keratin 32 |
| KRT37 | keratin 37 |
| LOH | Loss of Heterozygosity |
| MAP2K3 | mitogen-activated protein kinase kinase 3 |
| MAP3K4 | Mitogen-Activated Protein Kinase Kinase Kinase 4 |
| MAX | MYC Associated Factor X |
| MC1R | melanocortin 1 receptor |
| MDM2 | Mouse Double Minute 2 |
| MKI67 | marker of proliferation Ki-67 |
| MKS | Merkel Syndrome |
| MLLT1 | mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| MT1A | metallothionein 1A |
| mTOR | mammalian Target of Rapamycin |
| MUC20 | mucin 20, cell surface associated |
| MUC3A | mucin 3A |
| MYCN | v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| NEB | nebulin |
| NFAT | nuclear factor of activated T-cells |
| NONO | Non-POU Domain Containing Octamer Binding |
| NPHP | Nephronophthisis |
| NPIPB15 | nuclear pore complex interacting protein family, member B15 |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 |
| NRAP | nebulin-related anchoring protein |
| NRAP | nebulin-related anchoring protei |
| NWTSG | National Wilms’ Tumor Study Group |
| OBSCN | obscurin, |
| OR2B11 | olfactory receptor, family 2, subfamily B, member 11 |
| OR4C3 | olfactory receptor, family 4, subfamily C, member 3 |
| OR9G1 | olfactory receptor, family 9, subfamily G, member 1 |
| PCD | Primary Ciliary Dyskinesia |
| PCNT | pericentrin |
| PDE4DIP | phosphodiesterase 4D interacting protein |
| PDGFR | Platelet-derived growth factor |
| PHLPP2 | PH domain and leucine rich repeat protein phosphatase 2 |
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| PKD | Polycystic Kidney Disease |
| PKD1 | polycystic kidney disease 1 |
| PKD1L2 | polycystic kidney disease 1-like 2 |
| PKD2 | polycystic kidney disease 2 |
| PLD | phospholipase D family, |
| PLIN4 | perilipin 4 |
| PMFBP1 | polyamine modulated factor 1 binding protein 1 |
| POLYPHEN | Polymorphism Phenotyping |
| PPP3CA | protein phosphatase 3, catalytic subunit, alpha isozyme |
| PRIM2 | primase, DNA, polypeptide 2 |
| PRKAG2 | protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| PTCH1 | Patched 1 |
| PTCHD3 | patched domain containing 3 |
| PTPRH | protein tyrosine phosphatase, receptor type, H |
| PZP | pregnancy-zone protein |
| RHBG | Rh family, B glycoprotein |
| RNF43 | ring finger protein 43 |
| RP1L1 | retinitis pigmentosa 1-like 1 |
| RPGRIP1L | retinitis pigmentosa GTPase regulator interacting protein 1 like |
| SCGB1C1 | secretoglobin, family 1C, member 1 |
| SHh | Sonic Hedgehog |
| SIFT | Sort Intolerant From Tolerant |
| SIX1 | SIX homeobox 1 |
| SIX2 | SIX homeobox 2 |
| SMO | smoothened, frizzled class receptor |
| SNP | Single Nucleotide Polymorphism |
| SOX7 | SRY (sex determining region Y)-box 7 |
| STK3 | serine/threonine kinase 3 |
| TAF1C | TATA box binding protein (TBP)-associated factor |
| TARGET | Therapeutically Applicable Research to Generate Effective Treatments |
| TGF-B | Transforming Growth Factor-Beta |
| TMEM231 | transmembrane protein 231 |
| TP53 | tumor protein p53 |
| TRPC1 | transient receptor potential cation channel, subfamily C, member 1 |
| TRPV4 | transient receptor potential cation channel, subfamily V, member 4 |
| TUBB3 | tubulin, beta 3 class III |
| TZ | Transition Zone |
| UKCCSG | United Kingdom Children’s Cancer Study Group |
| VEP | Variant Effects Predictor |
| Wnt | Wingless/Integrated |
| WNT11 | wingless-type MMTV integration site family, member 11 |
| WT | Wilms Tumor |
| WT1 | Wilms tumor 1 |
| XPO5 | exportin 5 |
| ZCCHC14 | zinc finger, CCHC domain containing 14 |
| ZNF19 | zinc finger protein 19 |
| ZNF778 | zinc finger protein 778 |
AUTHOR CONTRIBUTIONS
E.K. was responsible for conducting the sequencing experiments and analysis of the microarray data to identify chromosome 16qloss/LOH samples. C-S. C. performed the bioinformatics analysis of the exome data, identified variants and performed analysis of the effects of the variants at the protein level. J.K.C. acted as an advisor as an expert in the field, provided samples and proof read the manuscript. L.H. designed the experiments, obtained the TARGET data, analyzed the pathways and wrote the manuscript.
CONFLICT OF INTEREST
There is no conflict of interests.

- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
- 12.
- 13.
- 14.
- 15.
- 16.
- 17.
- 18.
- 19.
- 20.
- 21.
- 22.
- 23.
- 24.
- 25.
- 26.
- 27.
- 28.
- 29.
- 30.
- 31.
- 32.
- 33.
- 34.
- 35.
- 36.
- 37.
- 38.
- 39.
- 40.
- 41.
- 42.
- 43.
- 44.
- 45.
Last Modified: 2020-10-06 09:34:48 EDT
