Genes & Cancer
Genomic profiling of DVL-1 and its nuclear role as a transcriptional regulator in triple negative breast cancer
Monica Sharma1, Isabel Castro-Piedras1, Austin Dwight Rodgers1 and Kevin Pruitt1
1 Immunology and Molecular Microbiology, Texas Tech University Health Sciences Center, Lubbock, TX, USA
Correspondence to: Kevin Pruitt, email: [email protected]
Keywords: Dishevelled (DVL); ChIP-Seq; transcription; histone modifications; breast cancer
Received: July 07, 2021
Accepted: September 24, 2021
Published: October 13, 2021
Copyright: © 2021 Sharma et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Dishevelled-1 (DVL-1) mediates Wnt signals critical for development and cellular homeostasis. DVL-1 is also linked with tumorigenesis, however its association with specific breast cancer (BC) subtypes and how it contributes to tumorigenicity remains poorly studied. Herein, using bioinformatics and genomics analyses, we investigate the role of DVL-1 in different molecular subtypes of BC. Our results demonstrate that DVL-1 is highly expressed in triple-negative BC compared to non-cancer tissues and associated with various clinical factors that may contribute to poor prognosis and survival rate.
Another critical knowledge gap which remains poorly investigated involves the role of DVL-1 in the nucleus. While the cytoplasmic role of DVL-1 as a signaling hub has been extensively studied, the nuclear role of DVL-1 remains virtually unexplored. Herein for the first time, we have performed ChIP-Seq analyses to identify genomic regions targeted and regulated by DVL-1, thus highlighting its potential role as a regulator of transcription. Furthermore, we observed that DVL-1 peaks co-localize with H3K27me3 and EZH2, a repressive epigenetic mark and a histone methyltransferase respectively. Overall, our findings emphasize the importance of DVL-1 with TNBC-related pathology and identified unexpected gene targets of DVL-1, that may help explain the complexity of aberrant Wnt signaling in cancer.
INTRODUCTION
Triple-negative breast cancer (TNBC) constitutes approximately 15–20% of breast cancers and is known for its aggressiveness and disproportionately higher rates of metastasis relative to other BC subtypes. It is also characterized by lack of estrogen receptors (ER), progesterone receptors (PR) and the absence of HER2 receptor overexpression on tumor cells, thereby limiting the use of available targeted therapies as a treatment option. Several reports suggest that oncogenic signaling mediated by the Wnt pathway is prominently associated with the TNBC-subtype [1]. Constitutive Wnt signaling is known to modulate the tumor microenvironment via fine tuning the crosstalk between malignant cells and infiltrating immune cells, making it an attractive therapeutic target in the treatment of TNBC [2]. DVL proteins provide a molecular hub through which Wnt signaling is relayed from upstream ligands and receptors to downstream effectors in order to control various cellular functions such as proliferation, stem cell renewal and migration [3]. DVL proteins have been classically associated with developmental disorders in a range of model organisms and with Robinow Syndrome in humans [3–5]. Emerging evidence suggests that DVL is also critical for breast tumorigenesis. Recently, our group showed that DVL-1 proteins are highly expressed in TNBC compared to non-cancer samples [6]. To further investigate its role, herein, we perform a comprehensive gene expression analysis of DVL-1 in different subtypes of breast cancer using the TCGA breast cancer datasets available in Tumor Immune Estimation Resource (TIMER), Human Protein Atlas (HPA), Breast Cancer Gen-ExMiner, UALCAN, and Kaplan Meier Plotter databases. Our results indicate that breast cancer tissues exhibit high levels of DVL-1 compared to normal counterparts. Moreover, we found DVL-1 to be significantly associated with various clinical risk factors for breast cancer patients such as cancer stage, Research Paper menopausal status, ER, PR, HER2 receptor and triple-negative status. Interestingly, we also observed that high expression of DVL-1 is associated with poor overall survival rate in TNBC patients.
Another mystery surrounding DVL is its function in different subcellular compartments. DVL has been studied primarily as a cytoplasmic scaffold which promotes either β-catenin stabilization or cell migration [7, 8]. However, seminal studies show that DVL proteins shuttle between the cytoplasm and nucleus, which is essential for Wnt/β-catenin signaling [9–11]. Moreover, our group was the first to report novel post-translational lysine acetylation of DVL proteins which acts as a regulatory switch to promote its nuclear localization [6]. Limited reports demonstrated that DVL binds to the promoters of Wnt target genes such as cMyc, BMP4, and cyclinD1 [11, 12]. Moreover, our previous study extended this binding repertoire and demonstrated that DVL proteins bind and regulates genes such as CYP19A1, a target not previously recognized as a Wnt target gene [13]. For a more comprehensive analysis of DVL nuclear function, we screened for novel DVL-1 target genes and discovered that DVL-1 binds to various genes involved in cancer biology and T-cell mediated immunity in TNBC models. Furthermore, analysis using TIMER database and gene expression studies indicates that high levels of DVL-1 are inversely correlated with target genes including infiltration of CD8+ and CD4+ T cells, highlighting a potential connection of DVL-1 in cancer immunomodulation.
Collectively, our findings provide additional insights into the contribution of DVL-1 in triple-negative breast tumorigenesis as well as suggests that nuclear DVL-1 could modulate tumor microenvironment by transcriptionally regulating genes involved in various cellular functions. Therefore, these findings propose DVL-1 as a promising prognostic biomarker for triple-negative tumors, a breast cancer subtype which urgently needs effective treatment options.
RESULTS
DVL-1 is highly expressed in various tumor types including breast cancer
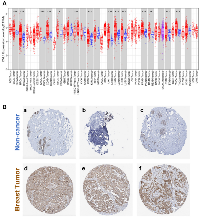
Dishevelled (DVL) proteins regulate oncogenic Wnt signaling which is a major driver of tumor progression [3]. Since DVL relays oncogenic input signals, we hypothesized that the expression levels of DVL-1 would be high in various cancer types. In order to understand the differential expression of DVL-1 between various tumor types (in red color) and its adjacent normal tissues (in blue color), we analyzed RNA-Seq data from TCGA. As shown in Figure 1A, using the TIMER database we found that DVL-1 was significantly upregulated in various cancer types which have hyper-activated Wnt signaling including breast cancer, colon adenocarcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, prostate adenocarcinoma compared to adjacent normal (healthy) tissues [14, 15]. In addition, to further validate the expression of DVL-1 protein in patient tissues, we used the pathology images from the HPA database, where DVL-1 is stained in normal and cancer tissues using antibody CAB011538. As presented in Figure 1B, the levels of DVL-1 antibody staining in three cases of breast cancer were high compared to the non-cancer adjacent tissues. Since the expression of both DVL-1 transcript and protein was significantly higher in breast tumor tissues, we next evaluated its association with different clinical parameters in breast cancer patients.
Association of DVL-1 expression with various clinical parameters and overall survival in breast cancer
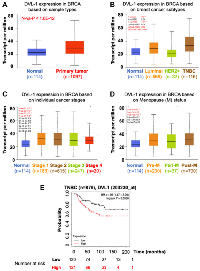
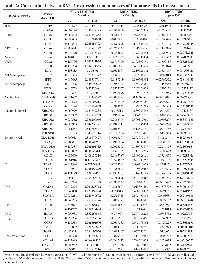
We assessed DVL-1 patterns in different molecular subtypes along with various clinical pathological parameters of breast cancer patients such as sample types, tumor stages, and menopausal status. Using TCGA breast cancer data cohort from UALCAN analysis platform, we confirmed that the expression of DVL-1 in breast cancer samples (n = 1097) was significantly higher than that in non-cancer/normal cancer tissues (n = 114) with a p-value <10−12 (Figure 2A). Next, the analysis based on different subtypes of breast cancer showed that expression of DVL-1 was significantly higher in luminal group (n = 566, p < 10−12) and triple-negative group (n = 116, p < 10−11) compared to normal control group. We observed that there was no statistical difference in the HER2+ subtype (n = 37, p < 0.24) of breast cancer compared to normal group. Interestingly, DVL-1 expression was significantly higher in triple negative subtype compared to other molecular subtypes including luminal (p < 0.02) and HER2+ (p < 0.006) groups (Figure 2B). Moreover, all stages of breast cancer had a significantly higher DVL-1 expression relative to the normal group. However, there was no significant difference in DVL-1 expression between the stages (Figure 2C). Expression of DVL-1 was also significantly higher in different classification of menopausal status: pre-menopausal (p < 10−11), peri-menopausal (p < 0.03) and post-menopausal status (p < 10−12) compared to normal (Figure 2D). Furthermore, we explored the correlation between DVL-1 mRNA levels and different risk factors such as lymph nodal status, ER status, PR status, HER2 status, and triple-negative status using a different database system called BC-GenExMiner 4.4 (Table 1). We observed that there was no difference in DVL-1 mRNA expression between nodal positive versus nodal negative status. However, based on the hormone receptor status, there was no significant different between ER-positive and ER-negative samples. However, DVL-1 mRNA levels were significantly highly expressed in PR-negative (p = 0.01), HER2-negative (p = 0.01) and triple-negative status (p = 0.004), further substantiating the idea that DVL-1 expression may serve as a potential diagnostic biomarker for specific subtypes of breast cancer such as triple-negative breast cancer.
Since the bioinformatics analyses showed promising results suggesting DVL’s association with various clinical risk factors for breast cancer, we wanted to explore the correlation between gene amplification of DVL-1 and patient survival. Using Kaplan Meier survival curves, we compared whether different expression levels of DVL-1 has any association with overall survival (OS) in triple-negative breast cancer patients. Notably, we observed that high DVL-1 was significantly associated with lower overall survival, hence poor prognosis in TNBC patients. The hazard ratio was significantly higher in TNBC group with a p-value equivalent to 0.009 (Figure 2E). Moreover, the results indicate that there was no significant association between DVL-1 expression and survival rates in luminal A/B, and HER2+ breast cancer subtypes (Supplementary Figure 1). These findings show that DVL-1 is significantly associated with mortality risk, suggesting that DVL-1 could serve as a prognostic marker for TNBC patients.
DVL-1 binds to different genomic locations in breast cancer models
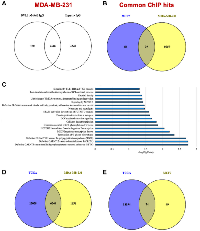
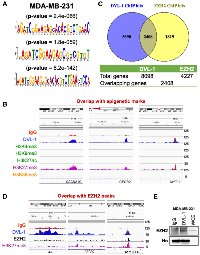
DVL plays a critical role in relaying Wnt signals within the cytoplasm for cellular growth, however its role in the nucleus remains virtually unexplored. Intrigued by the recent discoveries made by our group regarding the identification of a “regulatory switch” that controls nuclear translocation [6], we performed three independent DVL-1 ChIP-seq analyses in MDA-MB-231 and MCF7 cells to evaluate the role of nuclear DVL-1. For the ChIP-Seq experiments we used an IgG-control ChIP-Seq dataset as a reference for peak selection which showed a robust reproducibility between experiments in both cell lines. We obtained a total of 8098 peaks for DVL-1 in MDA-MB-231 cells (≥2-fold enrichment over IgG, intersecting 7348 Input genes), and a total of 123 peaks for DVL-1 in MCF7 (≥2-fold enrichment over IgG). Interestingly, 39 of the filtered MCF7 peaks overlapped with MDA-MB-231 peaks (Figure 3A–3B). Furthermore, we performed pathway analysis using the Reactome database which indicated that DVL-1 gene hits were involved in various biological processes including signaling transduction, post-translational modifications, protein-protein interactions, and human-associated diseases (Figure 3C). To investigate whether these findings could also be linked with breast cancer patients, we analyzed RNA-Seq data from 1904 breast cancer patients using cBioportal database (TCGA analysis). The RNA-Seq data was downloaded from Breast cancer study (Metabric, Nature 2012 & Nat Comm 2016), where about 18,268 genes were altered in primary breast tumors [16]. We found that there was an overlap of 6,240 genes and 74 genes altered in breast cancer patients and DVL-1 ChIP Seq hits in MDA-MB-231 and MCF7 cells respectively, which corresponds to 31% of DVL-1 hits in MDA-MB-231 and 0.4% of DVL-1 hits in MCF7 (Figure 3D–3E), further suggesting the importance of subtype-specific DVL-1 associations in patient tumors.
DVL-1 localizes at cancer-associated genes and regulates their expression
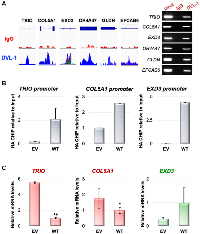
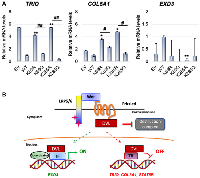
We previously reported that DVL proteins not only localize to the nucleus of breast cancer cells, but it also binds to CYP19A1 promoters and regulates its transcription [13]. Using the ChIP Seq bam files, we visualized the exact genomic location of DVL-1 binding (blue peak) at various cancer-associated genes not previously designated as Wnt target genes such as TRIO, COL5A1, EXD3, OR4A47, GLDN, and EFCAB6 compared to IgG control (red peak) using IGV (Figure 4A, Supplementary Figure 2). Next, we designed ChIP primers that spanned DVL-1 binding location on multiple genes and validated DVL occupancy by ChIP-PCR in MDA-MB-231 cells (Figure 4B). Additionally, we performed mRNA expression analysis to determine DVL-1 mediated regulation of target genes in MDA-MB-231 cells.Using RT-qPCR, we observed a reduction in the mRNA expression of TRIO and COL5A1 genes in WT (wild-type) MDA-MB-231 cells overexpressing DVL-1 compared to EV (empty vector) control cells. We tested the over-expression of DVL-1 on another gene, EXD3, however there does not appear to be a significant change in EXD3 expression with DVL-1 over-expression, suggesting a multidimensional role of DVL-1 that may depend on the genomic context (Figure 4C). These results clearly demonstrate that DVL-1 binds to multiple novel target genes and regulates the mRNA expression of some of its target genes we tested here.
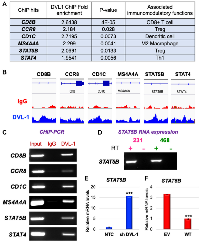
DVL-1 localizes to genes crucial for immune regulation and inversely regulates infiltrating T cell genes
In our ChIP-Seq screen, we discovered that DVL-1 localizes at genomic regions critical for immune regulation as listed in Figure 5A. We used IGV to visualize the exact genomic location of DVL-1 binding (blue peak) at different genes compared to IgG control (red peak) as presented in Figure 5B. We designed ChIP primers that spanned DVL-1 binding location on multiple lymphocyte genes and then validated DVL occupancy at CD8B, CCR8, CD1C, MS4A4A, STAT5B, and STAT4 genes by ChIP-PCR in MDA-MB-231 cells (Figure 5C, Supplementary Figure 3). Additionally, we performed mRNA expression analysis to determine the expression of DVL-1 targets in MDA-MB-231 cells. Interestingly, we observed that out of 6 genes only STAT5B mRNA was detected in TNBC cells, suggesting that DVL-1 may likely bind to transcriptionally repressed genes (Figure 5D). Furthermore, we wanted to evaluate whether DVL-1 regulates target genes such as STAT5B, which has been shown to play a critical role in cancer and anti-tumor immunity [17]. Using quantitative RT-PCR, we found that knockdown of DVL-1 resulted in upregulation of STAT5B expression, suggesting that DVL-1 acts as a repressor of STAT5B (Figure 5E). In contrast, over-expression of WT-DVL1 resulted in a significant reduction in STAT5B expression (Figure 5F), further validating that DVL-1 could potentially modulate anti-tumor immune responses by localizing to different genomic sites and inversely regulating expression of genes critical for anti-tumor immunity. To further understand the relationship between DVL-1 expression and different immune cell populations, we used the TIMER database to examine the correlation between DVL-1 expression and genes expressed by different lymphocyte subsets such as CD4+ T helper cells (e.g. Th1, Th2 and Th17), CD4+ regulatory T cells (Treg), follicular helper T cells (Tfh), CD8+ T cell and B cells. In addition, we used the TIMER database to determine the correlation between DVL-1 expression and genes expressed by different innate immune cells including monocytes, tumor-associated macrophages (TAM), M1/M2 macrophages, neutrophils, NK cells and dendritic cells in luminal, HER2-positive, and TNBC subtypes of breast cancer as summarized in Table 2 and Supplementary Figure 4. In our screen, we analyzed the immune cell genes that significantly correlated with DVL-1 expression with a p-value less than 0.05 (as highlighted in Table 2). We found that expression of DVL-1 was significantly correlated with immune cell genes in different breast cancer subtypes (Luminal, n = 611; HER2-positive, n = 67; and TNBC, n = 139). Particularly, the expression levels of DVL-1 was negatively and significantly correlated with 16 immune cell gene markers in TNBC breast cancer, such as CD2, CD86, CCL2, CD68, IL10, MS4A4A, KIR3DL2, HLA-DRA, HLA-DPA1, STAT4, STAT1, GATA3, IL21, CCR8, CTLA4, and HAVCR2. Interestingly, expression of some immune cell genes and DVL-1 uniquely correlated with luminal breast cancer, such as CD19, CD79A, CD8B, CCR7, CD1C, and STAT5B.
Interestingly, we found that DVL-1 mRNA expression is inversely correlated with B cell antigen (CD19), T-cell co-receptor (CD8A, CD8B), transcription factors that play a major role in innate and adaptive immune response (STAT1, STAT4), human leucocyte antigens (HLA) genes that are critical for encoding the major histocompatibility complex (MHC) II genes (HLA-DRA, HLA-DPA1), inflammatory cytokines (IFNG) and lymphocyte chemokine receptors (CCR7). Importantly, we found that the immunosuppressive cytokine, IL-10 was positively associated with DVL-1 mRNA expression in breast cancer patients. Taken together, these results suggest that there may be an important link between DVL-1 expression in breast cancer cells and the suppression of T cell mediated anti-tumor responses, perhaps by transcriptionally repressing the expression of various immune cell-related genes.
Nuclear DVL-1 binding is enriched on specific motif sequences and localizes with histone methyltransferase EZH2 and H3K27me3 epigenetic mark
To identify the DNA motifs driving the association of DVL-1 to the genome, we analyzed the genomic DNA sequences from DVL-1 binding peaks detected by ChIP-Seq analyses. All the peaks were submitted to MEME-ChIP analysis to determine significant motifs that had a strong central enrichment among the DNA sequences for the ChIPs performed. MDA-MB-231 motifs were: AGATCGGAAGAGCACACGTCTGAAC (p-value = 2.4e-066), AGATCGGAAGAGCGTCGTGT (p-value = 1.8e-059), AGATCGGAAGAGCACACGTAACTCCA (p-value = 8.2e-142) (Figure 6A).
The negative correlation between DVL-1 and expression of its direct target genes, suggested that DVL-1 may facilitate gene silencing at distinct genes, therefore we assessed whether DVL-1 can complex with repressive epigenetic marks. Thus, we evaluated DVL’s genomic localization with H3K27me3, an epigenetic modification associated with transcriptional repression. Visualizing the genomic regions of some of the top DVL-1 hits such as CACNA1C, CECR2, and MYT1L in MDA-MB-231 cells (Figure 6B), we observed that DVL-1 peaks overlapped with the epigenetic mark H3K27me3. In contrast, there was an absence of active epigenetic marks – H3K4me3, H3K9me3, H3K27ac, and H3K36me3 at DVL-1 binding regions, perhaps indicating that DVL-1 might be localizing at genes that are silenced in MDA-MB-231 cells.
Intrigued by the role of DVL-1 in the trimethylation of H3K27, a hallmark of gene repression, we asked whether DVL-1 may act as a scaffold of any H3K27 methyltransferase implicated in H3K27 methylation. In our screen we identified EZH2 as a good candidate and investigated further. Interestingly, our data shows that DVL-1 and EZH2 co-localize at several regulatory regions. Using Venny tools, we observed an overlap of 2408 genes between DVL-1 peaks with published EZH2 ChIP-Seq data (Figure 6C). Next, to determine if there is a link between DVL1-EZH2-H3K27me3, we first observed an overlap of the DVL-1 peaks with EZH2 at the genomic regions of our representative panel of novel DVL-1 targets such as ABR, CECR2, and MYT1L (Figure 6D). To further assess the significance of this finding, we also evaluated the physical interaction between DVL-1 and EZH2 by co-immunoprecipitation (Co-IP). Using Co-IP screen, we confirmed that EZH2 binds to DVL-1 in the MDA-MB-231 cell line (Figure 6E), indicating that DVL-1 and EZH2 may be part of a complex to regulate gene transcription. Altogether these data suggest that DVL-1 colocalizes with H3K27me3 and EZH2 at silenced regulatory regions of the chromatin.
Conserved lysines K69 and K285 on DVL-1 influences regulation of multiple target genes critical for tumorigenesis
Since K69 and K285 residues are known regulatory switches for DVL-1 nuclear localization [6], we wanted to determine whether these K-residues on DVL-1 also influence transcript levels of DVL-1 target genes such as TRIO, COL5A1 and EXD3 implicated in cancer. RNA was isolated from MDA-MB-231 cells stably expressing EV, WT-DVL-1, deacetylation mutants (K69R, K285R), as well as acetylation mutants (K69Q, K285Q). We used real-time qPCR with β-actin as an internal control to determine if WT versus mutants altered mRNA levels of several DVL-1 target genes that are critical for tumorigenesis. Interestingly, we found that WT, K69 and K285 acetylation mimetic (K69Q and K285Q) corresponded with a statistically significant decrease in the TRIO and COL5A1 transcripts with respect to EV and deacetylation mutants (K69R and K285R) in MDA-MB-231 cells. Furthermore, we observed that there was a significant increase in transcript levels of EXD3 in MDA-MB-231 cells with respect to K285R deacetylation mimetics (Figure 7A). These results show that nuclear DVL-1 and K-residues promoting its nuclear localization correlate with decreased gene expression, suggesting that nuclear DVL-1 is frequently linked with transcriptional repression of its target genes.
Altogether, we found that DVL-1 localizes at various transcriptional regulatory regions in breast cancer. As summarized in the model, DVL-1 binds to genomic regions and regulates transcription by potentially acting as a scaffold for transcription factors, suggesting a novel role of DVL-1 as a dynamic transcriptional regulator, both as an activator or repressor, based on the genomic location in the nucleus (Figure 7B).
DISCUSSION
DVL proteins are an integral hub of the Wnt signaling pathway, that mediates oncogenic Wnt signals to downstream to propagate tumor cell proliferation, differentiation, and migration. Wnt signaling is notoriously hyperactivated in different tumor types such as gastrointestinal cancer, leukemia and breast cancer [3, 15, 18]. Alteration in DVL proteins have been reported for several cancer types. For instance, a study using glioma as a model, suggested that DVL expression may contribute to the malignancy and invasion of human glioma [19]. Very recently, nuclear DVL proteins have been proposed to play an essential role in promoting canonical Wnt pathway. Accumulation of DVL-2 in the nucleus has been shown to result in hyper-activation of Wnt signaling in colorectal cancer [20]. Another study reported that nuclear DVL has been linked to lymph node metastases and poor prognosis of non-small-cell lung carcinoma [21]. DVL-1 nuclear staining was observed in almost 54% of cases with a significant increase in nuclear β-catenin staining in lung carcinomas [22]. Likewise, other studies suggested the carcinogenic potential of DVL proteins in chronic lymphocytic leukemia and breast cancer [23, 24]. Amplification of DVL-1 was observed in 13 of 24 primary breast cancer patients in comparision to corresponding non-cancerous breast tissues, with a significant association in nuclear localization of DVL and β-catenin proteins [25].
While several studies suggest that DVL proteins could serve as a potential prognostic biomarker for breast cancer and other cancer types, the true potential of DVL-1 and its contribution to clinical factors which regulate breast tumor progression and tumor immunology remained unanswered. This study utilizes publicly available data, bioinformatics tools, and genomic analyses to emphasize the contribution of DVL-1 in the most aggressive subtype of breast cancer known as triple-negative breast cancer. We start with examining the expression levels of DVL-1 in breast cancer using bioinformatics analysis which indicates high DVL-1 expression in breast cancer compared to non-cancer breast tissues, specifically in luminal B and triple-negative tumors. Moreover, we found a correlation between DVL-1 and several risk-factors for breast cancer using UALCAN and bc-Gen ExMiner databases. While assessing few clinical parameters, we found that DVL-1 expression was significantly associated with cancer stages, menopausal status, and triple-negative status in breast cancer patients. Herein, we extensively investigate association between DVL-1 expression with breast cancer subtypes, clinicopathological parameters, and various risk factors associated with breast cancer. Interestingly, we also found that high expression of DVL-1 was associated with poor overall survival in basal or triple-negative breast cancer (TNBC), suggesting that the expression of DVL-1 could affect the prognosis of triple-negative breast cancer. In that line, we then extended our efforts to predict the correlation between DVL-1 expression and its role in modulating gene expression and immune cell infiltration in TNBC tumors. Here, we would like to note that while TNBC is often used as a surrogate for basal breast cancer and there can be significant overlap in gene expression profiles, the two tumor types can be biologically distinct. TNBC tumors are defined by a negative immunohistochemical staining for ER, PR, and HER2 receptor, while the basal subtype is defined by a distinct gene expression marked by strong expression of markers such as cytokeratins 5, 6 and 17 [26].
Few reports have investigated DVL nuclear localization, and little is known about the scope of its binding to genomic locations. In previous studies, we not only demonstrated that DVL translocates to the nucleus across multiple breast cancer cell lines but also that DVL-1 binds to multiple aromatase tissue-specific promoters and regulates their transcription [13]. We also showed that increase in DVL-1 localization at breast/adipose promoter (I.4) promoter resulted in significant decrease in the levels of I.4 and total aromatase transcript [6]. This important finding led us to pursue of a new gene targets which were not previously associated with the Wnt pathway. Using ChIP sequencing analysis, we have identified that DVL-1 localizes at over 8000 genomic loci in breast cancer models. These genes hits were significantly associated with pathways/functions such as developmental biology, GPCR signal transduction, endocrinology, ion channels, and metabolism of proteins. Interestingly, we found DVL-1 binding at various genes which have an important role in tumorigenesis and breast cancer metastasis.
The immune system can be activated by tumor antigens, and when primed can elicit antitumor response that results in tumor destruction. T-cell priming against tumor antigens followed by infiltration of the anti-tumor T cells is an important step to kill the tumor cells which leads to tumor control. However, often this response is hampered by several factors that can directly influence the efficacy of the immune response. Herein, our findings suggest that DVL-1 may be one of the factors that help the tumor cells to escape anti-tumor immune responses thereby allowing for the proliferation and growth of malignant cells. Wnt pathway is an important regulator of development and homeostasis of mammalian cells. Emerging reports suggest that Wnt signaling regulates T cell development and activation, suggesting that Wnt signaling controls immune regulation in mammalian system [27–30]. In addition, recent reports suggest that Disheveled 1 knockout (Dvl1–/–) mice causes dysfunction of Paneth cells along with an increase in CD8+ T cells in the gastrointestinal (GI) tract [31]. Moreover, recent clinical trials suggest that immune checkpoints inhibitors have reduced efficacy in TNBC patients that undergo neoadjuvant chemotherapy. Hutchinson et al. illustrates that the anti-tumor immune activity within tumor decreases during transition from primary to metastatic disease. Furthermore, they showed that TNBC progression was accompanied with decreased proportion of stromal tumor-infiltrating lymphocytes (TILs) including B cell, CD4+ naive T cell, CD8+ T cells and cancer-associated fibroblasts in metastatic TNBCs [32]. Here, we shed light on an important, yet unexplored aspect of DVL-1 expression and its association with the presence or absence of immune cell levels in breast cancer. Using genomic analysis (ChIP-Seq) we demonstrate that DVL-1 localizes to genes of several important immune cell genes in TNBC model. Furthermore, mRNA expression studies, demonstrate a negative correlation between DVL-1 expression and the markers for T cell exhaustion (e.g. CTLA4, HAVCR2), CD8+ T cell (CD8B), tumor-associated macrophages (TAM), T helper cell, and dendritic cells. Recent reports suggest that Wnt signaling promotes tumorigenesis by modulating tumor microenvironment by fine tuning the transformed cell and infiltrating immune cells [2]. Our analysis suggests that increased DVL-1 expression correlates with decreased expression levels of several immune cell genes and anti-tumor immune cell signaling regulators such as STAT5B. In addition, using RNA-Seq data from BC patients we observed a significant negative correlation between DVL-1 expression and CD8+ T cells markers, cDC1-recruting chemokines, T cell and NK cell-recruiting chemokines. Thus, these results could provide some clues as to why immune checkpoint inhibitors may be ineffective for breast cancer treatment where the levels of onco-proteins such as DVL-1 are relatively high.
Based on the classical Wnt model, cytoplasmic DVL-1 channels Wnt signals which activates canonical and non-canonical branches. However, in our genomic studies we noticed that nuclear DVL-1 seems to act as a transcriptional repressor localizing with histone methyltransferase EZH2 and a repressive H3K27me3 epigenetic mark at its target genes. We were also curious to understand whether Wnt target genes overlap with DVL ChIP hits. Based on the comprehensive ChIP Seq studies that we performed for DVL-1 and DVL-3 in breast cancer cell lines [33], we observed that DVL proteins target non-canonical genes. To further elaborate, we observed that only 9 DVL-1 ChIP hits overlap with Wnt target genes, namely CD44, EGFR, FGF18, FGF4, LEF1, LGR5, MMP26, NOS2, and SALL4 genes. This might suggest a possibility that DVL proteins have functional implications beyond the classical Wnt model and also highlights the complexity of the signaling cascades in cancer context. Additionally, in the current study we report for the first time that DVL1 is associated with H3K27me3, a key repressive epigenetic mark that localizes to transcription regulatory regions, and we find an important overlap between DVL1 genomic occupancy and H3K27me3 peaks. Early studies by Kleer at al demonstrated that EZH2, a Polycomb Group (PcG) protein and transcriptional repressor involved in regulating cellular memory, was strongly associated with breast cancer aggressiveness. Overexpression of EZH2 in immortalized human mammary epithelial cell lines was shown to promote anchorage-independent growth and cell invasion [34]. Subsequent studies demonstrated that EZH2 overexpression led to aberrant terminal end bud architecture in the mammary gland and intraductal epithelial hyperplasia. In addition, EZH2 was found to physically interact with β-catenin and co-localize with β-catenin in human intraductal epithelial hyperplasia [35]. These early studies provided a link between EZH2 and β-catenin. The current study is the first to establish a link between EZH2 and DVL, a key regulator of both canonical and non-canonical Wnt signaling. While we have demonstrated a physical link and an overlap in occupancy of DVL1 and EZH2 at genomic loci, more questions remain to be addressed regarding the functional significance of their association. Additionally, others have reported that the Polycomb Repressive Complex 2 (PRC2), of which EZH2 is a part, establishes repressive H3K27me3 epigenetic signatures on genes involved in Wnt-governed intestinal homeostasis and Wnt-linked intestinal crypt dysfunction [36]. However, much more investigation is needed to decipher the functional significance of this convergence. Given these important links, we reason that DVL1 could be a key mediator that relays information initiated via Wnt ligands leading to epigenetic changes important for cellular and morphological phenotypes.
Collectively, our study demonstrates that high levels of DVL-1 is observed in breast cancer versus healthy tissue and is markedly associated with poor prognosis in triple-negative breast cancer. DVL-1 expression appears to negatively correlate with the presence of immune cells within the tumor microenvironment suggesting that DVL-1 could serve as suitable immune-modulating, prognostic biomarker for triple-negative breast cancer, a breast cancer type with unmet therapeutic needs.
MATERIALS AND METHODS
Tumor immune estimation resource data analysis
TIMER (Tumor IMmune Estimation Resource) is a web server for comprehensive analysis of tumor-infiltrating immune cells. Here we analyze the expression of DVL-1 in different cancer types, where tumor vs. normal sample types is plotted on x-axis and the DVL-1 gene expression is represented by log2 TPM. This resource is designed to systematically estimate tumor infiltrating cells such as B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells across diverse cancer types using a TIMER algorithm [37]. Moreover, this database can be used to explore the correlation between gene expression and abundance of immune infiltrates from RNA-seq expression profile. Correlation between DVL-1 and immune infiltration gene markers were explored using TIMER.
Human protein atlas analysis
HPA (https://www.proteinatlas.org) database aims to map all human proteins in different cell types, tissues and organs using various omics techniques like antibody-imaging and mass spectrometry-based proteomics [38]. This resource compared protein levels between normal and cancer tissues using immunostaining. In our study, we determine the expression of DVL-1 proteins between normal and breast cancer tissues under pathology section in HPA database (Antibody: CAB011538).
UALCAN analysis
UALCAN (http://ualcan.path.uab.edu) is a user-friendly web resource for analyzing OMICS data (TCGA and MET500) [39]. In this study, UALCAN was used to plot graphs depicting DVL-1 gene expression based on different clinical parameters in breast cancer carcinoma data set. The relative expression of DVL-1 was plotted based on parameters such as sample types (normal and primary tumor), breast cancer subclasses (normal, luminal, HER2 positive and TNBC), individual cancer stages (Stage 1, 2, 3, and 4), and menopausal status (normal, pre-menopause, peri-menopause, and post-menopause).
Kaplan-Meier plotter database analysis
The Kaplan Meier Database (https://kmplot.com/analysis) is used to evaluate effect of genes on survival status in different cancer types. We compare the expression of DVL-1 and survival status in different molecular subtypes of breast cancer including luminal A (n = 2504), luminal B (n = 1425), HER2+ (n = 335), and TNBC group (n = 879). Overall survival (OS) were evaluated using Kaplan Meier plotter database. Hazard ratio (HR) for 95% confidence interval and significance value (p) less than 0.05 were considered statistically significant.
Breast cancer GenExMiner 4.4 analysis
BC-Gen Ex Miner v4.4 (http://bcgenex.centregauducheau.fr/) statistical tool contains breast cancer (BC) transcriptome data (DNA microarrays and RNA-seq data) which allows gene expression analysis in breast cancer [40, 41]. This database contains three modules: correlation, expression, and prognosis. The correlation module identifies whether the gene-of-interest is either positively or negatively correlated with co-expressed genes located on the same chromosomal region. The expression analysis compares expression of genes to critical clinical parameters such as HER2, ER, PR status and other parameters. Lastly, the prognostic module allows the assessment of the prognostic impact of a gene on all possible combinations of population. For our study, we logged to the BC-Gen Ex Miner homepage, and selected RNA seq data and entered the DVL-1 as query gene. We then analyzed DVL-1 expression level based on various pre-determined clinical parameters such as nodal status (N+ positive or N− negative), estrogen receptor (ER+/ER−), progesterone receptor (PR+/PR−), HER2 status (HER2+/ HER2−), triple-negative status (TNBC and not-TNBC). The data is summarized as Table 1, where p-value less than 0.05 were considered statistically significant.
Cell culture
All cell lines (MDA-MB-231, and MCF7) used in this manuscript were purchased from ATCC which utilizes STR technology for Cell Authentication, and they were used in a low passage (<20) within 6 months or less after receipt or resuscitation. MDA-MB-231 cells were cultured in DMEM (Gibco, 11995-065), while MCF7 cells were cultured in MEM (Gibco, 11095-080) supplemented with 0.1% insulin (Sigma, I0516). All culture media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco, 15070-063).
Chromatin immunoprecipitation (ChIP), ChIP-Seq and data analysis
ChIP was done according to a published protocol [6, 13]. Briefly, cells were grown to confluence in 100 mm dishes; a final count of approximately 5 × 106 cells per plate. Cells were cross-linked with 1% (w/v) formaldehyde for 8 min at room temperature and quenched in 0.125 M of glycine for 5 minutes at room temperature. The medium was removed, and cells were washed twice with PBS. Cells were scraped, pelleted and washed twice with PBS plus protease inhibitor cocktail. Cells were resuspended in SDS Lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM 0.5 M EDTA, and 1% SDS) with protease inhibitor cocktail. Cells were then sonicated in a Diagenode Bioruptor sonicator for 30 cycles (30 sec pulses and 30 sec rest) and washed twice with chilled 1X PBS containing a protease inhibitor cocktail, scraped off the plates and collected by centrifugation. The soluble chromatin fraction was quantitated and 100 μg of chromatin was immunoprecipitated with anti-DVL1 antibody (D3570; Sigma), or Rabbit IgG (I5006; Sigma) for 2 h at 4°C. Dynabeads Protein A (Invitrogen, 10002D) were added to the chromatin-antibody mixture and incubated with rotation for 2.5 hours at 4°C. ChIPs were washed with five low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris HCl pH 8.1, and 150 mM NaCl), three high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris HCl pH 8.1, and 500 mM NaCl), and one TE wash (1 mM EDTA and 10 mM Tris HCl pH 8). Immunoprecipitated chromatin samples were reverse-crosslinked overnight at 65°C, followed by RNAse A (Promega) at 37°C for 2 hours, and proteinase K incubation (Promega) at 55°C for 2 hours. DNA was eluted using Qiaquick PCR purification kit (Qiagen) and amplified by end-point PCR and real time qPCR using gene-specific primers (Table 3A). DNA from ChIP was extracted, and at least 10 ng ChIP DNA was sent to GENEWIZ and used for ChIP-Seq library preparation and sequencing. FASTQ files were analyzed using DNASTAR’s Laser Gene software. Sequenced reads were aligned against the human genome and the peaks were visualized using integrative genomic viewer (IGV). MEME-ChIP was used to analyze DVL-1 binding motifs and TOMTOM to identify if those motifs were similar to known consensus sequences using the MEME Suite Programs http://meme-suite.org/index.html [42]. Venn diagrams to identify the overlapping genes were generated using the Venny tool [43].
The Cancer Genome Atlas (TCGA) data analysis
Genomic data from Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) study was analyzed using cBio Cancer Genomics Portal (http://cbioportal.org). The study consisted of gene expression analysis from 1904 breast cancer patient [44]. Correlation analysis was conducted to determine genes which were common between genes altered in breast cancer patients and DVL-1 ChIP-Seq analyses in MDA-MB-231 and MCF7 cells.
Stable expression of WT and mutants in MDA-MB-231 cell lines
350,000 cells were plated in a p6-well dish. After 24 hours, cells were transfected with 1 µg DNA of empty vector (pcDNA3.1(+) without HA-tag), full-length DVL-1 (WT), K to R, K to Q point-mutants using lipofectamine 3000 reagent (Thermo Scientific). After 24 hours, the cells were selected using 1 mg/ml G418 antibiotic until no cells were remaining in non-transfected control.
DVL-1 stable knock-down
MDA-MB-231 were infected by pLKO.1-puro based shRNA MISSION lentiviral transduction particles for DVL-1 (TRCN0000441114, Sigma), and Non-Target shRNA control transduction particles (SHC002V, Sigma). 24 h prior to transduction, cells were plated in order to reach 80% confluency at the time of transduction. The transduction was enhanced with Hexadimethrine Bromide (Sigma, H9268) at a final concentration of 8 μg/ml. Following the addition of hexadimethrine bromide, the appropriate amount of viral particles were added at 2× multiplicity of infection (MOI) to the media, which was replaced with fresh media after 24 h. The puromycin selection was started 48 h after transduction at a concentration 1 μg/ml of puromycin (Gibco, A11138-03), and the puromycin-containing media was replaced every 3–4 days until total selection was achieved.
mRNA expression analysis
Total RNA was isolated from cells using a Bio-Rad RNA extraction kit. 2 μg of total RNA was reverse-transcribed using SuperScriptIII Reverse Transcriptase (ThermoFisher) to synthesize first-strand of complementary DNA (cDNA). Intron-spanning primers were designed for each specific target DNA and gene expression measured by either endpoint-PCR or real-time qRT-PCR (Table 3B).
End-point PCR and quantitative real-time qRT-PCR
End-point PCR amplification was performed using JumpStart RedTaq (Sigma). The Applied Biosystems Veriti 96-well thermal cycler (Applied Biosystems) and Gel DOC EZ imager (Bio-Rad) were used analyses. Gene expression was quantified by real-time qPCR in CFX96 Normal-Well Real-Time System (BioRad) using PerfeCta SYBR Green FastMix and specific oligonucleotide primers. The reaction mixtures contained 10 µl PerfeCta SYBR Green FastMix, 7.2 µl ddH2O, 2.0 µl template cDNA and 0.4 µl gene-specific 10 µM PCR oligonucleotides primers. The reaction conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s and Melt Curve (dissociation stage). Relative gene expression was calculated as delta (Δ Re (the difference between the cycle threshold values, Ct, of the internal control, and Ct of gene of interest) and confirmed by 2–ΔΔ CT method.
Co-immunoprecipitation
Cells were seeded and incubated at 37°C under conventional O2 conditions. Once they were 70% confluent, cells were washed with PBS and lysed in Co-IP lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol and Protease inhibitor cocktail). This was followed by quantification for equal protein loading using standard BCA protocol (Thermo Scientific, 23227). Cell extracts were incubated with 2 μg of DVL-1 specific antibody (D3570; Sigma), and species-matched IgG as a negative control for 2 hours/4°C. Protein A Dynabeads (Life Technologies) were incubated with the antigen-antibody complex for 2 hours at 4°C. Beads were washed four times with lysis buffer with gentle agitation for 5 minutes per wash. 5× sample buffer (Invitrogen) was used for elution of complex from beads followed by Western blotting along with WCE.
Immunoblots
Samples were subjected to polyacrylamide gel electrophoresis using Invitrogen Nupage gel system, transferred to PVDF (Millipore) membranes, and immunoblotted. Antibodies used are as follows: EZH2 (cst-5246; Cell Signaling). Membranes were incubated in 5% BSA/TBST with primary antibody overnight at 4°C. Membranes were washed with TBST and probed with horseradish peroxidase-conjugated secondary antibodies in 5% BSA/TBST for 1 hour/room temperature (RT). Membranes were visualization by enhanced chemiluminescence (ECL) reagent (Thermo Scientific) using Azure C300 gel imaging system (Azure Biosystems).
Statistical analysis
The difference in expression of DVL-1 gene in different samples types, cancer subclasses, and clinical parameters were analyzed using UALCAN and BC-GenExMiner database. Moreover, the difference between high and low expression of DVL-1 in survival curves were obtained using Kaplan-Meier method. The results from these databases were with p-value and log-hazard ratios (later for survival curves only) from a log-rank test. The Pearson correlation coefficient was used to measure any correlation among genes, where 0 = |r| represents no correlation, 0.0 < |r| < 0.2 is weak correlation, 0.2 ≤ |r| < 0.4 is low moderate correlation, 0.4 ≤ |r| < 0.6 is moderate correlation, 0.6 ≤ |r| ≤ 0.8 is strong correlation, 0.8 ≤ |r| < 1.0 represents very strong correlation, and 1.0 = |r| : perfect correlation. The same is true for negative correlation. Lastly, p-value (p) less than 0.05 was considered statistically significant.
Author contributions
MS and KP discussed and designed the experiments. MS, ICP, ADR performed experiments and data analysis for ChIP-Seq experiments. MS wrote and edited the paper with input from all authors. MS, ICP, ADR, and KP reviewed and approved the manuscript.
ACKNOWLEDGMENTS AND FUNDING
This research was supported by the National Institute of Health (CA155223) to K.P. and a Cancer Prevention and Research Institute of Texas (CPRIT), Recruitment of Rising Stars Award (RR410008) to K.P.
CONFLICTS OF INTEREST
Authors have no conflicts of interest to declare.

- 1. Wnt signaling in triple-negative breast cancer. Oncogenesis. 2017; 6:e310. https://doi.org/10.1038/oncsis.2017.14. [Pubmed]
- 2. Wnt Signaling and Its Significance Within the Tumor Microenvironment: Novel Therapeutic Insights. Front Immunol. 2019; 10:2872. https://doi.org/10.3389/fimmu.2019.02872. [Pubmed]
- 3. Dishevelled: A masterful conductor of complex Wnt signals. Cell Signal. 2018; 47:52–64. https://doi.org/10.1016/j.cellsig.2018.03.004. [Pubmed]
- 4. The Dishevelled Protein Family: Still Rather a Mystery After Over 20 Years of Molecular Studies. Curr Top Dev Biol. 2016; 117:75–91. https://doi.org/10.1016/bs.ctdb.2015.11.027. [Pubmed]
- 5. Dishevelled Paralogs in Vertebrate Development: Redundant or Distinct? Front Cell Dev Biol. 2017; 5:59. https://doi.org/10.3389/fcell.2017.00059. [Pubmed]
- 6. Acetylation of conserved DVL-1 lysines regulates its nuclear translocation and binding to gene promoters in triple-negative breast cancer. Sci Rep. 2019; 9:16257. https://doi.org/10.1038/s41598-019-52723-3. [Pubmed]
- 7. Multiprotein complexes governing Wnt signal transduction. Curr Opin Cell Biol. 2018; 51:42–49. https://doi.org/10.1016/j.ceb.2017.10.008. [Pubmed]
- 8. Dishevelled: The hub of Wnt signaling. Cell Signal. 2010; 22:717–27. https://doi.org/10.1016/j.cellsig.2009.11.021. [Pubmed]
- 9. The sirtuins promote Dishevelled-1 scaffolding of TIAM1, Rac activation and cell migration. Oncogene. 2015; 34:188–98. https://doi.org/10.1038/onc.2013.549. [Pubmed]
- 10. Nuclear localization is required for Dishevelled function in Wnt/beta-catenin signaling. J Biol. 2005; 4:3. https://doi.org/10.1186/jbiol20. [Pubmed]
- 11. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J Cell Biol. 2008; 180:1087–100. https://doi.org/10.1083/jcb.200710050. [Pubmed]
- 12. Frizzled 7 expression is positively regulated by SIRT1 and beta-catenin in breast cancer cells. PLoS One. 2014; 9:e98861. https://doi.org/10.1371/journal.pone.0098861. [Pubmed]
- 13. DVL1 and DVL3 differentially localize to CYP19A1 promoters and regulate aromatase mRNA in breast cancer cells. Oncotarget. 2018; 9:35639–54. https://doi.org/10.18632/oncotarget.26257. [Pubmed]
- 14. Wnt Pathway: An Integral Hub for Developmental and Oncogenic Signaling Networks. Int J Mol Sci. 2020; 21:8018. https://doi.org/10.3390/ijms21218018. [Pubmed]
- 15. Wnt signaling in cancer. Oncogene. 2017; 36:1461–73. https://doi.org/10.1038/onc.2016.304. [Pubmed]
- 16. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun. 2016; 7:11479. https://doi.org/10.1038/ncomms11479. [Pubmed]
- 17. STAT5 in Cancer and Immunity. J Interferon Cytokine Res. 2016; 36:226–37. https://doi.org/10.1089/jir.2015.0054. [Pubmed]
- 18. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012; 4:a008052. https://doi.org/10.1101/cshperspect.a008052. [Pubmed]
- 19. The expression and significance of dishevelled in human glioma. J Surg Res. 2014; 192:509–14. https://doi.org/10.1016/j.jss.2014.06.034. [Pubmed]
- 20. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest. 2017; 127:3375–91. https://doi.org/10.1172/JCI94292. [Pubmed]
- 21. Coexpression of IQ-domain GTPase-activating protein 1 (IQGAP1) and Dishevelled (Dvl) is correlated with poor prognosis in nonsmall cell lung cancer. PLoS One. 2014; 9:e113713. https://doi.org/10.1371/journal.pone.0113713. [Pubmed]
- 22. Brain metastases from lung cancer show increased expression of DVL1, DVL3 and beta-catenin and down-regulation of E-cadherin. Int J Mol Sci. 2014; 15:10635–51. https://doi.org/10.3390/ijms150610635. [Pubmed]
- 23. Dishevelled proteins are significantly upregulated in chronic lymphocytic leukaemia. Tumour Biol. 2016; 37:11947–57. https://doi.org/10.1007/s13277-016-5039-5. [Pubmed]
- 24. Amplification, up-regulation and over-expression of DVL1, the human counterpart of the Drosophila disheveled gene, in primary breast cancers. Cancer Sci. 2003; 94:515–18. https://doi.org/10.1111/j.1349-7006.2003.tb01475.x. [Pubmed]
- 25. Wnt signaling pathway in invasive ductal carcinoma of the breast: relationship between beta-catenin, dishevelled and cyclin D1 expression. Oncology. 2007; 73:112–17. https://doi.org/10.1159/000120999. [Pubmed]
- 26. Basal-like and triple-negative breast cancers: searching for positives among many negatives. Surg Oncol Clin N Am. 2014; 23:567–77. [Pubmed]
- 27. Signaling pathways governing stem-cell fate. Blood. 2008; 111:492–503. https://doi.org/10.1182/blood-2007-07-075168. [Pubmed]
- 28. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998; 8:11–20. https://doi.org/10.1016/s1074-7613(00)80454-9. [Pubmed]
- 29. Wnt signaling is required for thymocyte development and activates Tcf-1 mediated transcription. Eur J Immunol. 2001; 31:285–93. https://doi.org/10.1002/1521-4141(200101)31:1%3C285::AIDIMMU285%3E3.0.CO;2-D. [Pubmed]
- 30. Beta-catenin stabilization extends regulatory T cell survival and induces anergy in nonregulatory T cells. Nat Med. 2008; 14:162–69. https://doi.org/10.1038/nm1707. [Pubmed]
- 31. Dual epithelial and immune cell function of Dvl1 regulates gut microbiota composition and intestinal homeostasis. JCI Insight. 2016; 1:e85395. https://doi.org/10.1172/jci.insight.85395. [Pubmed]
- 32. Comprehensive Profiling of Poor-Risk Paired Primary and Recurrent Triple-Negative Breast Cancers Reveals Immune Phenotype Shifts. Clin Cancer Res. 2020; 26:657–68. https://doi.org/10.1158/1078-0432.CCR-19-1773. [Pubmed]
- 33. Nuclear Dishevelled targets gene regulatory regions and promotes tumor growth. EMBO Rep. 2021; 22:e50600. https://doi.org/10.15252/embr.202050600. [Pubmed]
- 34. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003; 100:11606–11. https://doi.org/10.1073/pnas.1933744100. [Pubmed]
- 35. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009; 175:1246–54. https://doi.org/10.2353/ajpath.2009.090042. [Pubmed]
- 36. Polycomb Repressive Complex 2 Enacts Wnt Signaling in Intestinal Homeostasis and Contributes to the Instigation of Stemness in Diseases Entailing Epithelial Hyperplasia or Neoplasia. Stem Cells. 2017; 35:445–57. https://doi.org/10.1002/stem.2479. [Pubmed]
- 37. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017; 77:e108–10. https://doi.org/10.1158/0008-5472.CAN-170307. [Pubmed]
- 38. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018; 27:233–44. https://doi.org/10.1002/pro.3307. [Pubmed]
- 39. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017; 19:649–58. https://doi.org/10.1016/j.neo.2017.05.002. [Pubmed]
- 40. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford). 2013; 2013:bas060. https://doi.org/10.1093/database/bas060. [Pubmed]
- 41. bc-GenExMiner: an easyto-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012; 131:765–75. https://doi.org/10.1007/s10549-011-1457-7. [Pubmed]
- 42. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–08. https://doi.org/10.1093/nar/gkp335. [Pubmed]
- 43. VENNY: An interactive tool for comparing lists with Venn Diagrams. 2007. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 44. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–04. https://doi.org/10.1158/2159-8290.CD-12-0095. [Pubmed]
