Genes & Cancer
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
Daniela S. Bassères1, Aaron Ebbs2, Patricia C. Cogswell2 and Albert S. Baldwin2,3
1Department of Biochemistry, Chemistry Institute, University of São Paulo, São Paulo, SP, Brazil;
2Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC;
3Department of Biology, University of North Carolina, Chapel Hill, NC.
Correspondence to: Albert S. Baldwin, email: [email protected]
Keywords: Lung cancer, KRAS, NF-κB, IKK, p53
Received: July 2, 2013
Accepted: April 21, 2014
Published: April 21, 2014
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT:
Activating mutations in KRAS are prevalent in cancer, but therapies targeted to oncogenic RAS have been ineffective to date. These results argue that targeting downstream effectors of RAS will be an alternative route for blocking RAS-driven oncogenic pathways. We and others have shown that oncogenic RAS activates the NF-κ B transcription factor pathway and that KRAS-induced lung tumorigenesis is suppressed by expression of a degradation-resistant form of the IκBα inhibitor or by genetic deletion of IKKβ or the RELA/p65 subunit of NF-κB. Here, genetic and pharmacological approaches were utilized to inactivate IKK in human primary lung epithelial cells transformed by KRAS, as well as KRAS mutant lung cancer cell lines. Administration of the highly specific IKKβ inhibitor Compound A (CmpdA) led to NF-κB inhibition in different KRAS mutant lung cells and siRNA-mediated knockdown of IKKα or IKKβ reduced activity of the NF-κB canonical pathway. Next, we determined that both IKKα and IKKβ contribute to oncogenic properties of KRAS mutant lung cells, particularly when p53 activity is disrupted. Based on these results, CmpdA was tested for potential therapeutic intervention in the Kras-induced lung cancer mouse model (LSL-KrasG12D) combined with loss of p53 (LSL-KrasG12D/p53fl/fl). CmpdA treatment was well tolerated and mice treated with this IKKβ inhibitor presented smaller and lower grade tumors than mice treated with placebo. Additionally, IKKβ inhibition reduced inflammation and angiogenesis. These results support the concept of targeting IKK as a therapeutic approach for oncogenic RAS-driven tumors with altered p53 activity.
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths worldwide [1] and even though novel targeted therapies have been developed that show efficacy for a subset of patients [2,3], for the great majority of lung cancer patients effective targeted therapies are still lacking. This is the case for the 20-50% of lung cancer patients that harbor activating point mutations in the KRAS GTPase gene [4-6]. Therefore, identification of druggable targets in the KRAS signaling pathway could lead to novel therapeutic alternatives for lung cancer, as well as other RAS-driven cancers.
Constitutive, signal-independent activation of KRAS via mutation is not only associated with poor prognosis and therapy resistance in a variety of cancers, but is sufficient to trigger malignant transformation and drive the oncogenic phenotype [7,8]. Therefore, KRAS is a rational target for cancer therapy. Unfortunately, due to the difficulty in effectively inhibiting the biological activity of RAS proteins, approaches to directly target these proteins for therapy have been so far unsuccessful [9]. In this regard, intense efforts are being made to target known downstream effectors of RAS [10,11,12]. So far this approach has yielded limited therapeutic success, thus reflecting the need to better understand the molecular pathways triggered by oncogenic RAS.
A mechanism that is known to be important for RAS-induced oncogenesis is the activation of the transcription factor NF-κB. NF-κB is a ubiquitously expressed transcription factor that is maintained in an inactive form through interactions with the inhibitor of κB (IκB) proteins. Canonical NF-κB activation downstream of inflammatory cytokines and other inducing molecules is mediated by the IκB kinase (IKK) complex, which is comprised of a regulatory subunit (NEMO) and two catalytic subunits (IKKα and IKKβ). Once activated, the IKK complex phosphorylates IκB, which leads to its rapid ubiquitination and proteasome-mediated degradation. In this pathway, the p50-p65/RELA heterdodimer is then released and accumulates in the nucleus to regulate target gene transcription [13-17]. In the non-canonical NF-κB pathway, NIK activates an IKKα homodimer to lead to nuclear accumulation of a p52-RELB heterodimer [13,14,16,17]. Additionally, IKKε and TBK1, IKK-related kinases can activate p65- and c-REL-containing complexes [18,19].
We previously demonstrated that NF-κB is activated downstream of oncogenic RAS and that inhibition of NF-κB leads to RAS-induced cell death [20,21]. Inhibition of NF-κB by expression of the super-repressor form of IκBα [22] or deletion of the RELA/p65 subunit of NF-κB [23] blocks KRAS-induced lung tumors. In that latter work, we demonstrated that KRAS activates the transcription factor NF-κB in lung tumors in situ and that loss of p65 in the tumors leads to the induction of apoptosis [23]. Barbie et al [24] have shown that the IKK-related kinase TBK1 is important as a survival factor in KRAS-driven cancer cells, potentially through a mechanism that involves c-REL. Duran et al [25] demonstrated that oncogenic KRAS can activate IKK through the signaling adaptor p62 and other studies have shown that genetic deletion of IKKβ in different cancer models suppresses RAS-induced tumorigenesis [26-28].
Here we show that pharmacological inhibition of IKKβ in primary human lung epithelial cells transformed by KRAS, as well as KRAS mutant lung cancer cell lines, inhibits NF-κB activity and reduces cell growth. Further analysis indicated that this response was at the level of cellular proliferation and not induction of cell death. Genetic targeting of KRAS, IKKβ or IKKα by siRNA had similar effects on NF-κB activity, reducing canonical NF-κB activation. In addition, cell growth and proliferation were also similarly affected. Nonetheless, even though NF-κB activity was reduced in all cells examined, reduced cell growth was restricted to cells with lost or disrupted p53 function. Therefore, we treated a KRAS-induced lung cancer mouse model combined with loss of the tumor suppressor p53 with a highly specific IKKβ inhibitor (Compound A, Bayer [29]). The inhibitor is well tolerated and lowers tumor burden and tumor grade. Consistent with the cell-based studies, Compound A (CmpdA) treatment reduces tumor proliferation. CmpdA also affects the tumor microenvironment, reducing the tumor-associated macrophage footprint along with reduced intratumoral vasculature. These results show that IKKα or IKKβ inhibition reduces lung cancer cell proliferation in vitro and pharmacological IKKβ targeting reduces lung cancer growth in vivo, supporting the hypothesis that IKK inhibition therapy will have clinical benefits in lung cancer as well as other cancers, particularly for patients with KRAS mutations and altered p53 activity.
RESULTS
IKK targeting decreases canonical NF-κB activity in KRAS mutant cell lines
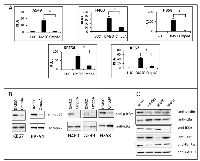
We had previously shown that oncogenic KRAS expression in low-passage primary immortalized human small airway cells correlates with increased IκB phosphorylation and NF-κB DNA binding [23]. In addition, IKK inhibition with CmpdA reduced NF-κB DNA binding activity in these genetically defined cells [23]. To further address the contribution of IKK to oncogenic RAS-driven NF-κB activity, human as well as mouse lung cancer cell lines harboring oncogenic KRAS were transfected with an NF-κB-dependent luciferase reporter and treated with CmpdA. In each experiment, the IKK inhibitor reduced NF-κB reporter activity (Fig. 1A). CmpdA also blocked phosphorylation of IKKβ substrates IκBα or p65 in these cell lines (Fig. 1B). Furthermore, when KRAS or IKKβ expression was reduced by transfecting human lung cancer cell lines with siRNA targeting either KRAS or IKKβ, NF-κB luciferase reporter activity was also inhibited, indicating that, in these cells, both KRAS and IKKβ are promoting NF-κB activity (Supplementary Fig. S1).
Based on the results showing IKKβ is involved in KRAS-induced NF-κB activation, and based on published evidence that NF-κB activation by KRAS in lung cells involves the IKK complex [23,25], we hypothesized that in human lung cells KRAS utilizes the canonical pathway to activate NF-κB. In this regard, we questioned whether siRNA-mediated inhibition of IKKα would also affect NF-κB activity in KRAS mutant lung cells. IKKα is typically considered to be less important in canonical NF-κB signaling as compared to IKKβ [30]. Interestingly, knockdown of IKKα in the lung cancer cells studied not only reduced NF-κB activity (Supplementary Fig. S1), but more importantly, inhibition of KRAS, IKKβ or IKKα by siRNA in H358 cells inhibited IκBα phosphorylation and degradation, a hallmark of the canonical NF-κB activation pathway (Fig. 1C).
IKK targeting reduces proliferation of KRAS positive cells dependent on the loss of p53 function
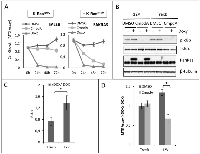
Next, we examined the effects of CmpdA treatment on growth of several KRAS positive cells. Interestingly, in spite of reducing NF-κB activity in all cell lines studied, CmpdA did not affect cell growth uniformly. SAKRAS cells are very sensitive to CmpdA treatment, whereas their isogenic cells lacking oncogenic KRAS (SALEB) are less sensitive (Fig. 2A), indicating that the effect of CmpdA on cell growth is dependent on KRAS status. In order to further assess this dependency, we used H1703 lung adenocarcinoma cells, which harbor wild-type KRAS, to generate HA-tagged KRASG12V-inducible H1703 human lung cancer cells. We observed that induction of KRASG12V expression with doxycycline in H1703 cells leads to increased IκBα phosphorylation and to increased NF-κB reporter activity (Figs. 2B and 2C). These effects on NF-κB activity were not observed in doxycycline-treated empty vector control cells (H1703-TrexB cells), which do not express KRASG12V (Figs. 2B and 2C). CmpdA treatment blocked IκBα phosphorylation in both KRASG12V and TrexB H1703 cells (Fig. 2B). We then used this cell line model to evaluate CmpdA sensitivity. Induction of KRASG12V expression with doxycycline in H1703 cells leads to enhanced cell growth, which is blocked by treatment with CmpdA (Fig. 2D). CmpdA had no effect on growth of H1703 TrexB cells, which do not express KRASG12V upon doxycycline administration (Fig. 2D).CmpdA sensitivity was also dependent on the status of the tumor suppressor p53. KRAS positive cell lines with wild-type (WT) p53 (A549 and H460) were less sensitive to CmpdA treatment than KRAS positive p53 null (H358) or p53 mutant (H1792) cell lines (Fig. 3A). In order to address a role for p53 in the sensitivity to IKK inhibition, we analyzed cells isolated from KRAS-induced lung cancer mouse models with WT p53 (KE67) and with deletion of the p53 gene (KPF54). Results demonstrate that the tumor cells with loss of p53 are more sensitive to IKKβ inhibition than cells with WT p53 (Fig. 3A).
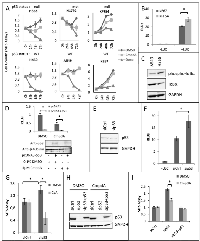
Because loss of p53 can increase IKKβ activity [31] and has been shown to activate NF-κB in KRAS-transformed lung tumor cells [22], we propose that, even though NF-κB is active in all KRAS positive cell lines, loss or disruption of p53 activity would to lead to enhanced IKKβ activation and thus to enhanced CmpdA sensitivity (see discussion). In order to determine if p53 null cells have higher NF-κB activity than p53 WT cells, we performed luciferase reporter assays on murine cells derived from KRAS-induced lung tumors with either WT p53 or with loss of p53. As can be seen in Fig. 3B, KPF54 cells, which lack p53, display higher NF-κB reporter activity than KE67 cells, which express WT p53. Similar results were observed when we compared NF-κB activity between human cells with different p53 status. A549 cells, which express WT p53 have lower levels of phosphorylated IκBα (Fig. 3C), a hallmark of NF-κB activation. Finally, we expressed p53 in human H358 cells, which are p53 null, and this resulted in reduced NF-κB reporter activity (Fig. 3D).
In order to determine if the increased sensitivity to CmpdA displayed by p53 null cell lines can be caused by loss of p53, we used RNA interference to reduce p53 expression in p53 WT KE67 murine cells and in p53 WT A549 human cells. A 58% reduction in p53 expression in KE67 cells (Fig. 3E) results, not only in increased NF-κB reporter activity (Fig. 3F), but also in increased cell growth, which is reduced by CmpdA treatment (Fig. 3G). Interestingly CmpdA had no effect on KE67 cells transfected with a control small interfering RNA (siRNA). Similar results were observed in A549 cells. Inhibition of p53 expression resulted in increased A549 cell growth and rendered A549 cells sensitive to CmpdA treatment (Figs. 3H and 3I). More importantly, p53 re-expression in these cells, not only reduced cell growth, but turned them insensitive to IKKβ inhibition (Figs. 3H and 3I). In order to further corroborate this data we used HCT116 colon cancer cells that harbor oncogenic KRAS and that have been engineered by homologous recombination to lose the p53 gene, making it straightforward to evaluate the effect of p53 loss in an isogenic setting. Not only is NF-κB activity higher in HCT116-p53 knockout (KO) cells than in HCT116-p53 WT cells (Supplementary Fig. S2A and S2B), but also, as assessed by MTT assays, HCT116-p53 WT cells are insensitive to CmpdA treatment, whereas HCT116-p53 KO cells are sensitive (Supplementary Fig. S2C). When we used clonogenic cell growth assays, both cell lines were sensitive to CmpdA treatment, but they displayed different levels of sensitivity. Whereas HCT166-p53 WT cells showed a 55% reduction in colony formation, HCT116-p53 KO cells showed an 80% reduction (Supplementary Fig. S2D). Taken together these results indicate that the status of both KRAS and the p53 tumor suppressor are important in determining sensitivity to IKKβ inhibition therapy.
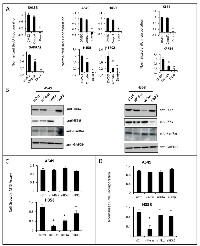
In order to determine the mechanism leading to reduced cell growth induced by CmpdA, we analyzed both apoptosis and reduced proliferation. Interestingly, even though NF-κB is known as an antiapoptotic transcription factor, growth reduction was not caused by apoptotic cell death, as CmpdA failed to induce apoptosis in any of the cell lines (Supplementary Fig. S3A). However, reduction in cell growth in all sensitive cell lines was associated with reduced proliferation as measured by BrdU incorporation (Fig. 4A).
In order to validate IKKβ as the relevant CmpdA target in mediating reduced proliferation, we pursued siRNA experiments in H358 cells and A549 cells (Fig. 4B). The A549 cell line, which is resistant to CmpdA treatment, is also resistant to alterations in cell growth (Fig. 4C) and proliferation (Fig. 4D) upon IKKβ knockdown. On the other hand the H358 cell line, which is sensitive to CmpdA treatment, is more sensitive to loss of IKKβ by siRNA, displaying a 27% reduction in cell growth (Fig. 4C) and 20% in proliferation (Fig. 4D). The reduction in proliferation observed upon IKKβ knockdown is not as robust as that attained with CmpdA treatment (20% versus 39%). This could be caused by residual expression of IKKβ in the knockdown experiments and/or a compensatory increase in IKKα activation, which could affect cell proliferation (see below). siRNA-mediated knockdown of KRAS in H358 cells led to a greater reduction of cell growth (Figs. 4C and 4D), suggesting that KRAS activates additional pathways that contribute to cell proliferation.
Because IKKα is a member of the IKK complex and because siRNA targeting of IKKα in KRAS positive cells also reduces NF-κB activity and inhibits IκBα phosphorylation/degradation (Supplementary Fig. S1 and Fig. 1C), it is important to determine the contribution of this kinase to the proliferative phenotype of KRAS positive/p53 null lung cancer cells. Therefore, we determined if siRNA knockdown of IKKα would affect cell growth, apoptosis and proliferation. Interestingly, like inhibition of IKKβ, siRNA to IKKα in p53 WT A549 cells did not affect cell growth or proliferation, whereas in the CmpdA sensitive H358 cell line knockdown of IKKα also decreases cell growth and proliferation (Fig. 4C and 4D), suggesting that both IKKα and IKKβ contribute to promote cell proliferation. Again, consistent with the observed lack of effect of CmpdA on apoptosis, knockdown of IKKα, IKKβ or even KRAS in these cell lines, did not trigger apoptosis (Supplementary Fig. S3B).
To further corroborate these results, we examined a panel of apoptosis and proliferation-related genes in H358 cells following knockdown of KRAS, IKKα or IKKβ. Whereas, no significant changes were observed in the expression of anti-apoptotic genes BCL2 and CIAP2, expression of the proliferation-related genes E2F1 and MYC were affected (Supplementary Fig. S3C). Knockdown of KRAS inhibited expression of both E2F1 and MYC. Interestingly, even though to a lesser extent, knockdown of either IKKα or IKKβ also inhibited E2F1
Last Modified: 2016-06-14 13:25:16 EDT
PII: 5
