Genes & Cancer
Screening of suppressors of bax-induced cell death identifies glycerophosphate oxidase-1 as a mediator of debcl-induced apoptosis in Drosophila
Jessie Colin1,2,*, Julie Garibal1,2,*, Amandine Clavier1,2, Sébastien Szuplewski1, Yanick Risler1, Cécile Milet1,2, Sébastien Gaumer1, Isabelle Guénal1,3 and Bernard Mignotte1,2,3
1 Université Versailles St-Quentin, Laboratoire de Génétique et Biologie Cellulaire, Montigny-le-Bretonneux, France.
2 Ecole Pratique des Hautes Etudes, Laboratoire de Génétique Moléculaire et Physiologique, Montigny-le-Bretonneux, France
3 Co-senior authors
* These two authors contributed equally to this work
Correspondence to: Bernard Mignotte, email: [email protected]
Keywords: Apoptosis, Bax, Debcl, mutagenesis, Glycerophosphate oxidase
Received: February 9, 2015
Accepted: June 6, 2015
Published: June 10, 2015
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Members of the Bcl-2 family are key elements of the apoptotic machinery. In mammals, this multigenic family contains about twenty members, which either promote or inhibit apoptosis. We have previously shown that the mammalian pro-apoptotic Bcl-2 family member Bax is very efficient in inducing apoptosis in Drosophila, allowing the study of bax-induced cell death in a genetic animal model. We report here the results of the screening of a P[UAS]-element insertion library performed to identify gene products that modify the phenotypes induced by the expression of baxin Drosophila melanogaster. We isolated 17 putative modifiers involved in various function or process: the ubiquitin/proteasome pathway; cell growth, proliferation and death; pathfinding and cell adhesion; secretion and extracellular signaling; metabolism and oxidative stress. Most of these suppressors also inhibit debcl-induced phenotypes, suggesting that the activities of both proteins can be modulated in part by common signaling or metabolic pathways. Among these suppressors, Glycerophosphate oxidase-1 is found to participate in debcl-induced apoptosis by increasing mitochondrial reactive oxygen species accumulation.
INTRODUCTION
Major executioners of programmed cell death by apoptosis are relatively well conserved throughout evolution. However, the control of commitment to apoptosis exhibits some differences between organisms. During mammalian cells apoptosis, various key pro‑apoptotic factors are released from the inter‑membrane space of mitochondria (for review, see [1]). These factors include cytochrome c, Apoptosis Inducing Factor (AIF), Endonuclease G, Smac/DIABLO (Second mitochondria‑ derived activator of caspase/direct IAP‑binding protein with low PI) and the serine protease Omi/HtrA2. Once released in the cytosol, cytochrome c binds to the WD40 domain of Apaf‑1 and leads to the formation of a cytochrome c/Apaf‑1/caspase‑9 complex called “apoptosome”, in which caspase‑9 (a cysteinyl aspartase) auto‑activates to initiate a caspase activation cascade that will lead to cell death. Mitochondrial permeabilization is under the control of the Bcl‑2 family of proteins. These proteins share one to four homology domains with Bcl‑2 (named BH1‑4) and exhibit very similar tertiary structures. However, while some of these proteins (such as Bcl‑2) are anti‑apoptotic, the others are pro‑apoptotic and assigned to one of the following sub‑classes: BH3‑only proteins (such as Bid) and multi‑domain proteins (such as Bax). During apoptosis, Bax translocates to the mitochondrial outer membrane, undergoes conformational changes, oligomerizes and finally allows the release of pro-apoptotic factors from the intermembrane space (review [2]). Anti‑apoptotic proteins of the Bcl‑2 family oppose this Bax‑mediated mitochondrial release of apoptogenic factors while BH3‑only proteins can activate Bax or inhibit anti‑apoptotic proteins of the family.
In C. elegans, activation of the caspase CED‑ 3 requires CED‑4, the homologue of Apaf‑1 but no cytochrome c. The Bcl‑2 family protein CED‑9 constitutively interacts with CED‑4 and thereby prevents the activation CED‑3. This repression of cell death is released upon binding of CED‑9 to the BH3‑only protein EGL‑1, which induces a conformational change in CED‑9 that results in the dissociation of the CED‑4 dimer from CED‑9. Released CED‑4 dimers form tetramers, which facilitate auto‑activation of CED‑3 [3]. Although CED‑9 appears bound to mitochondria, these organelles seem to play a minor role in apoptosis in C. elegans, contrarily to mammals [4].
The role of mitochondria in Drosophila programmed cell death remains more elusive [1, 5‑7]. Cytochrome c does not seem crucial in the apoptosome activation [8, 9], which is mediated by the degradation of the caspase inhibitor DIAP1 by proteins of the Reaper/Hid/Grim (RHG) family. The apoptotic cascade appears somehow inverted between flies and worm/mammals. In these two last organisms, apoptosis regulators are relocated from mitochondria to the cytosol. Contrarily, Drosophila apoptosis regulators are concentrated at or around mitochondria during apoptosis. Indeed, targeting the RHG proteins Reaper (Rpr) and Grim to mitochondria seems to be required for their pro‑apoptotic activity [10‑ 12]. Furthermore, Hid possesses a mitochondrial targeting sequence and is required for Rpr recruitment to the mitochondrial membrane and for efficient induction of cell death in vivo [13].
The important role played in Drosophila by the mitochondria in apoptosis is also suggested by the mitochondrial subcellular localization of Buffy and Debcl, the only two members of the Bcl-2 family identified, so far, in this organism. Buffy was originally described as an anti‑ apoptotic Bcl‑2 family member [14, 15], but it can also promote cell death [16‑19]. Debcl (death executioner Bcl 2 homolog), is a multidomain death inducer [19‑23] that can be inhibited by direct physical interaction with Buffy [14]. When overexpressed in mammalian cells, debcl induces both cytochrome c release from mitochondria and apoptosis. This protein interacts physically with anti‑ apoptotic members of the Bcl‑2 family, such as Bcl‑2 itself, in mammals. In Drosophila, Debcl is involved in the control of some developmental cell death processes as well as in irradiation‑induced apoptosis [15, 18, 24].
We have previously shown in Drosophila that mammalian Bcl‑2 inhibits developmental and irradiation‑ induced cell death [25] as well as rpr‑ and bax‑induced mitochondrial membrane potential collapse [26]. Interestingly, we have shown that bax‑induced cell death is mitigated by loss‑of‑function (LOF) mutations in genes encoding some components of the TOM complex which controls protein insertion in the outer mitochondrial membrane [5]. These results suggest that Bax mitochondrial location remains important for its activity in Drosophila. Therefore, flies provide a good animal model system to study Bax‑induced cell death in a simple genetic background and look for new regulators of Bcl‑2 family members.
Here, we report the results of the screening of P[UAS]‑element insertion (UYi) library, performed in order to identify modifiers of bax‑induced phenotypes in Drosophila. Among 1475 UYi lines screened, 17 putative modifiers were isolated, that include genes involved in various cellular functions. We also present a more detailed study of one of these modifiers, UY1039, and show that glycerophosphate oxidase‑1 (Gpo‑1) [EC 1.1.5.3] participates in debcl‑induced apoptosis by increasing reactive oxygen species (ROS) production.
RESULTS
A modifier screen for suppressors of bax-induced phenotypes
We have previously shown that expression of the proapoptotic gene bax induced apoptosis in the developing eye or wing [25, 26]. Expression of bax under control of the wing specific vg-GAL4 driver during development led to lethality and notches in the wing of the surviving escapers. As expected from the temperature sensitivity of the UAS‑GAL4 expression system, lethality was more penetrant and wing phenotypes were more severe when flies were raised at 25°C than at 18°C. This adult wing phenotype was suppressed by bcl-2 expression [25, 26] and by heterozygosity for LOF mutations in genes encoding Tom22 or Tom70 [5], indicating that the vg>bax-induced adult wing phenotype is sensitive and thus amenable to genetic screening.
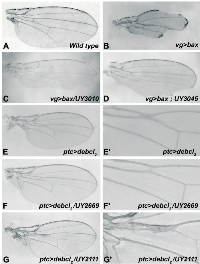
To gain insight into the molecular mechanism of bax‑induced apoptosis and with the aim of isolating regulators of this process, we designed a genetic screen for modifiers of Bax-mediated tissue loss in the wing. To ease the screening procedure, we used a strain recombined for vg-GAL4 and UAS-bax transgenes. Animals heterozygous for vg>bax showed a strong and scorable notched wing phenotype (Figure 1B, compared to 1A), facilitating the selection of suppressors rather than enhancers.
A mutagenesis involving the transposition of a P‑element containing UAS sequences, P[Mae-UAS.6.11] has been performed. Genes flanking the 5’ end of the transposon could therefore be transcriptionally regulated by the UAS/GAL4 system and thus overexpressed. The P‑element insertion, per se, could also generate LOF mutations. As part of a consortium of laboratories, we have produced Drosophila lines with random insertion sites of the P‑element named hereafter UYi [27‑30]. Thus, a collection of 1475 lines, i.e. 594 with the P‑element inserted on the second chromosome, 775 on the third chromosome and 106 on the X chromosome was generated. Males carrying UYi were crossed with vg>bax/CyOGFP females and their progeny were screened for a rescue of both lethality (at 18°C and/or 25°C) and the notch phenotype (see Materials and Methods). Examples of wing phenotypes suppression are shown on Figure 1C and D. 56 UYi lines were selected, corresponding to 3,8% of the collection (56/1475).
Since the number of flies exhibiting a rescue of the wing phenotype was sometimes low, we tested the relevance of the suppression of the wing phenotype by a statistical approach. As previously described [26], expressivity and penetrance of the wing phenotype are variable in a population of flies of the same genotype. Flies expressing bax in the wing exhibit a distribution of phenotypes that can be classified into three categories: strong, intermediate and weak according to the number and size of notches observed along the wing margin. Therefore, we have used the statistical Wilcoxon test [31] to compare distributions of phenotypes between the two different types of progenies that express bax with or without the UYi suppressor. This test defines an α and a Ws value that respectively allow assessment of whether two distributions are significantly different or not, and which population is composed of stronger phenotypes. We defined the statistically significant limit as α<10‑3. Using this stringent criterion, we identified 24 suppressors of bax acting both on fly survival and wing phenotype, among the 56 selected UYi lines, corresponding to 1.6% of the collection (24/1475).
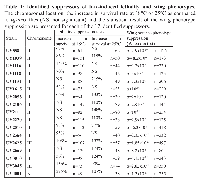
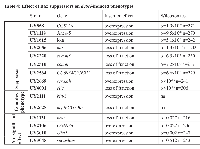
Subsequently, to distinguish possible additive effects from more specific interactions, all selected UYi lines were crossed with vg-GAL4. Only wild type wing phenotypes were observed, ruling out an additive effect of UYi and bax transgenes, and thus revealing the specificity of the genetic interactions between bax and the 24 insertions. To define if the wing phenotype suppression could be due to a gain of function (GOF) or a LOF, we identified the insertion point and orientation of each of the 24 insertions. PCR rescue experiments were performed and their products were sequenced. We compared the recovered sequence of the flanking genomic DNA to the Drosophila full‑genome sequence database [32]. We were unable to obtain unambiguous flanking sequence information for 7 isolated UYi lines, i.e. UY504, UY1220, UY1236, UY1649, UY2303, UY2650 and UY2803. Therefore, 17 putative suppressors of bax‑induced apoptosis were identified. The results obtained during the screen of these bax‑induced phenotypes are presented in table 1. Table 2 summarizes the genetic and molecular characterization of the selected insertions.
Candidate genes were regrouped according to the known or putative molecular function of their predicted products (Table 3). They are involved in cell growth, proliferation or death, pathfinding and cell adhesion, secretion and extracellular signaling, metabolism and oxidative stress, ubiquitin/proteasome pathway.
Study of the effects of bax suppressors on debcl-induced phenotypes
Bax seems to induce cell death in the eye thanks to its interaction with Debcl [24]. Moreover, only few data are reported concerning Debcl regulation and its partners. Therefore, we decided to test if the identified modifiers of bax‑induced cell death could also genetically interact with debcl.
As for bax‑induced cell death, the expression of debcl in wing imaginal discs during development induces apoptosis, which leads to a wing phenotype [14]. A very homogenous adult phenotype was obtained when expressing debcl along the antero‑posterior frontier of wing discs, thanks to ptc-GAL4 driver [33]. Under these conditions, debcl expression brought closer L3 and L4 veins in the proximal region of the wing, inducing a fusion of these veins in the region of the anterior cross vein (figure 1E and E’). These phenotypes were due to an excess of apoptosis [33].
Before testing genetic interactions, all selected UYi insertion were crossed with ptc-GAL4 flies, to verify that their own overexpression did not give rise to any wing phenotype. All lines studied showed a wild type wing phenotype when tested with ptc-GAL4. An exception was UY2669 that exhibited an absence of anterior cross vein but no alteration at the level of the antero‑posterior frontier.
This secondary screen was performed on 15 insertions because two lines were lost (UY1116 and UY1039). We found nine suppressors of debcl‑induced phenotype (Table 4). An example is shown in Figure 1F‑ F’ for UY2669. Four of Bax modifiers had no significant effect and two led to complex phenotypes of partial penetrance as shown for UY2111 in Figure 1G- G’. Thus, most of bax‑induced suppressors also suppressed debcl‑ induced apoptosis.
Glycerophosphate oxidase-1 participates in superoxide production during Debcl-induced apoptosis
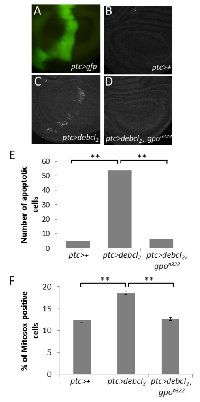
The line UY1039 being lost we assumed that the suppressor effect of UY1039 was due to a LOF mutation in the Glycerophosphate oxidase-1 gene. Given that this gene is involved in the mitochondrial metabolism, and that numerous Bcl‑2 family members act at the mitochondrial level, Gpo‑1 seemed of high interest and we decided to focus on its study. To confirm our hypothesis, we tested whether RNAi against Gpo-1 or heterozygosity for a Gpo-1 hypomorph (Gpo-1291) or an amorph (Gpo-1n322) allele, could suppress Debcl overexpression‑induced wing phenotype. Both RNAi and both Gpo-1 LOF heterozygous alleles suppressed debcl‑induced phenotypes but the most complete and fully penetrant suppression was observed in flies heterozygous for the Gpo-1n322 mutation (Figure 2). Therefore, we decided to assess the apoptosis level in wing imaginal discs of flies heterozygous for Gpo-1n322. The number of TUNEL positive cells in wing discs overexpressing debcl was dramatically reduced by Gpo-1n322 heterozygosity when compared to discs that are not mutated in Gpo-1 (Figure 3A-E), thus confirming that a reduction of Gpo-1 dosage suppresses debcl‑induced apoptosis.
Gpo‑1 [EC 1.1.5.3], also known as glycerol‑3‑ phosphate dehydrogenase, is encoded by a nuclear gene and located at the outer surface of the inner mitochondrial membrane. It catalyzes the reaction:
sn‑glycerol 3‑phosphate + coenzyme Q10 ‑> glycerone phosphate + reduced coenzyme Q10
Along with the cytosolic NAD-linked glycerol phosphate dehydrogenase (GPDH), Gpo‑ 1 forms the glycerol phosphate shuttle that catalyzes the interconversion of glycerol phosphate and dihydroxyacetone phosphate to oxidize cytosolic NADH by transferring reducing equivalents from the cytosol to mitochondria. Cytosolic GPDH has been shown to protect mammalian CHO cells against ROS‑induced apoptosis [34]. On the opposite, mitochondrial Gpo‑1 has been involved in superoxide production in various species (review: [35]) including Drosophila [36], which is in agreement with a protective role of a dosage reduction of this gene. Furthermore, we have observed that debcl overexpression can induce ROS accumulation thanks to a mitochondrial superoxyde indicator, i.e. MitoSOX. Indeed, debcl overexpression increases the proportion of MitoSOX positive cells in the wing imaginal discs compared to the ptc-gal4/+ control (Figure 3F). Thus, we decided to test whether heterozygosity for the Gpo-1 null mutant could decrease ROS levels produced by debcl overexpression. A significant decrease of MitoSOX positive cells was observed when debcl was overexpressed in a Gpo1n322 heterozygous background when compared to Gpo‑1+/+ background (Figure 3F). This result indicates that Gpo-1 participates in debcl‑induced apoptosis by increasing ROS production.
DISCUSSION
This screen provides us with 17 suppressors of phenotypes induced by the expression of bax under control of the wing specific vg-GAL4 driver (lethality and wing notches). The possibility that these suppressors affect GAL4 synthesis or that the selected insertions titrate the GAL4 transcription factor is unlikely, since our number of suppressors is limited (1.6% of the collection). Moreover, we isolated UYi insertions, which were not identified in other screens performed using the same collection and the UAS/Gal4 system [28, 30]. Finally, we have recently reported the specificity of one of the suppressors, UY3010, which corresponds to a gain‑of‑function of the Ubiquitin activating enzyme‑encoding gene Uba1. Indeed, Uba1 overexpression allows the degradation of Bax and Debcl, thanks to the activation of the ubiquitin/proteasome pathway [33]. We also showed in this study that Debcl is targeted to the proteasome by the E3 ubiquitin ligase Slimb, the β-TrCP homologue [33].
We found that 9 of the bax-modifiers also behaved as suppressors of debcl‑induced wing phenotype while 4 showed no significant effect on this phenotype. Three hypotheses could explain this discrepancy. One possibility is that these bax modifiers are context artifacts and do not represent bona fide Bax interactors. The second possible explanation involves the difference in the driver used in each assay (vg-GAL versus ptc-GAL). Indeed, UY3010 did not significantly suppress debcl‑induced apoptosis while another Uba1 overexpression mutant (Uba1EP2375) did [33]. Third, although Bax and Debcl, share similarities in their mode of action and regulation, some signaling pathways could be specific of bax‑induced apoptosis. Indeed, a LOF of brat mitigates neither debcl‑ (this paper) nor hid‑ or Sca3‑induced cell death [37].
The brat gene belongs to a group of suppressors, which is implicated in cell growth, proliferation or death. Mutations in this type of genes could compensate cell loss due to ectopic apoptosis induction. Results observed for this group of modifiers can generally be easily interpreted with the literature data. UY1131 corresponds to an insertion in the brat (for brain tumor) gene that could allow the expression of a truncated form of the protein. To check whether this insertion leads to a LOF or a GOF of brat, we tested the effect of the characterized LOF allele bratk0602 on bax‑induced phenotypes. This mutation strongly suppressed (α=7.3x10‑7) the wing phenotype showing that UY1131 is a LOF of brat (data not shown). Brat belongs to the NHL family of proteins, represses translation of specific mRNAs 38] and is a negative regulator of cell growth [39‑41]. The suppression of bax‑ induced phenotypes by a LOF of brat could suggest that this gene also regulates cell death, which seems unlikely according to its inability to suppress other cell death pathways [37]. Alternatively brat could regulate somehow compensatory proliferation in this system.
Some candidate suppressors encode proteins involved in secretion or components of the extra‑cellular matrix. The effect of these genes could rely on cell signaling. Change in levels of secreted proteins could modify cell‑extracellular matrix interactions and thus affect viability via processes similar to anoikis.
Several suppressors are implicated in pathfinding (comm, comm3, hat, scratch and lola). Two hypotheses can be formulated. Either neurons are of particular importance in bax‑induced phenotypes or a more general role of these proteins in signaling is responsible for these suppressions. If the neuronal death could explain the decreased survival of bax expressing flies, it could hardly explain the wing phenotypes. Therefore, these suppressor genes may have a more general role in signaling and in particular in cell death regulation. For example, UY2669 corresponds to a GOF mutant of scratch (scrt). This gene is a Drosophila homologue of C. elegans ces-1, which encodes a snail family zinc finger protein involved in controlling programmed death of specific neurons [42]. Interestingly, a mammalian homologue of scratch, named Slug, is involved in a survival pathway that protects hematopoietic progenitors from apoptosis after DNA damage [43]. Slug also antagonizes p53‑mediated apoptosis by repressing the bcl-2‑family pro‑apoptotic gene puma [44]. More recently, a regulatory loop linking p53/Puma with Scratch has been described in the vertebrate nervous system, not only controlling cell death in response to damage but also during normal embryonic development [45].
Another possibility is that these modifiers could affect some extracellular survival and/or death factors. For example, sugarless, which was found twice in the screen, has been shown to interact with several survival pathways such as Wingless, EGF and FGF pathways that can play a role in defining shape and size of tissues and organs. This result can be paralleled with the suppressive effect of mutations in hephaestus and lola, both of which interact with the Notch/Delta signaling. Notably, lola, a gene encoding a Polycomb group epigenetic silencer, has been shown to be required for programmed cell death in the Drosophila ovary [46]. Lola has also been identified for its role in normal phagocytosis of bacteria in Drosophila S2 cells and as a component of the Drosophila Imd pathway that is key to immunity [47]. In contrast, Lola is required for axon growth and guidance in the Drosophila embryo [48]. This indicates that lola could play a role in cell adhesion and motility. Accordingly, when coupled with overexpression of Delta, misregulation of pipsqueak and lola induces the formation of metastatic tumors associated with a downregulation of the Rbf (Retinoblastoma-family) gene [49].
Other identified genes are involved in carbohydrate metabolism (Gpo-1 and CG8036 described as a transketolase). This result is in agreement with the evidence that Bcl‑2 family proteins, in addition to their well characterized function in cell death, also play roles in metabolic processes in particular at the level of energetic metabolism (reviewed in [50]). In particular, Bcl‑2 regulates mitochondrial respiration and the level of different ROS through a control of cytochrome c oxidase activity [51]. Study of heterologous bax expression in yeast has provided clues on Bax function in relation to ROS (reviewed in [52, 53]) and yeast LOF mutants of genes involved in oxidative phosphorylation show increased sensitivity to Bax cytotoxicity [54]. In agreement, Bcl‑xL complements Saccharomyces cerevisiae genes that facilitate the switch from glycolytic to oxidative metabolism [55]. Furthermore, both the anti‑ apoptotic effect of LOF mutations in Gpo-1 and the GOF in transketolase genes can be related to a protective effect against oxidative stress. This result suggests that the cell death process induced by Bax involves, at least in part, the modulation of different ROS levels.
Indeed, we report here that the suppressor effect of a null allele of Gpo-1 is associated with a decreased ability of Debcl to induce ROS production. This result is in agreement with the observation that 70% of the total cellular H2 O2 production was estimated to stem from Gpo‑ 1 in isolated Drosophila mitochondria [36]. This enzyme has also been implicated in ROS production in mammalian brown adipose tissue mitochondria when glycerol‑3‑ phosphate was used as the respiratory substrate [56] and, more recently, in prostate cancer cells [57]. In this latter case, ROS production seems to be beneficial to cancer cells, whereas we show here that it favors cell death in Drosophila wing disc cells. This apparent contradiction could be related to the abnormal ROS production occurring during the oncogenic transformation and the shift to a glycolytic metabolism.
In conclusion, this study shows that Gpo‑1 contributes to debcl‑induced apoptosis by increasing reactive oxygen species (ROS) production and provides a substantial resource that will aid our efforts to understand the regulation of pro‑apoptotic members of the Bcl‑2 family proteins.
MATERIALS AND METHODS
Fly stocks
All strains were raised on standard culture medium at 25°C or 18°C. Generation of UYi lines was performed by standard P mobilization [58] of a P[Mae-UAS.6.11] [27]. The driver strains used in this study are vestigial-GAL4 (vg-GAL4) [25] and patched-GAL4 (ptc-GAL4) given by L. Théodore. The strain carrying UAS-bax (from mouse origin) has been generated in our laboratory and was previously described [25]. The strain carrying two insertions of UAS-debcl-HA (one on the second and one on the third chromosome) was given by H. Richardson [20] and recombined with the ptc-GAL4 driver to generate the pct>debcl2 strain. Either y,wc or w1118 Canton S was used as the control strain according to the genetic background of the tested lines. The Uba1EP2375 mutant strain was obtained from the Szeged Drosophila stock center. Gpo-1n322 [59], Gpo-1291 and UAS-RNAi-Gpo-1 Trip #55319 strains were obtained from the Bloomington Drosophila Stock Center. The UAS-RNAi-Gpo-1 VDRC KK #110608 strain was obtained from Vienna Drosophila RNAi Center.
Screen for suppressors of lethality
UYi lines including a P‑element insertion on the X, second or third chromosome were tested. Concerning the screening of UYi lines carrying a P‑element on the second chromosome, the survival rate in the progeny was evaluated as follows. Each cross was performed with 7 virgin females and 3 males. [vg>bax/CyOGFP] virgin female flies were mated to [UYi/UYi] males when homozygous males were viable or, alternatively, to heterozygous [UYi/CyO] males. In the first case, the survival rate corresponds to ([vg>bax/UYi] / [UYi/CyOGFP]) while in the case of heterozygous mutant strains it corresponds to (([vg>bax/CyO] + [vg>bax/UYi]) / [UYi/CyOGFP]).
For UYi lines carrying the P‑element on the third chromosome, virgin female flies [vg>bax/CyOGFP ; +/+] were mated to viable [+/+ ; UYi/UYi] males or, alternatively, to [+/+ ; UYi/TM3] males. In the first case, the survival ratio corresponds to ([vg>bax/+ ; UYi/+] / [CyOGFP/+ ; UYi/+]), while in the second case it was (([vg>bax/+ ; UYi /+] + [vg>bax/+ ; TM3/+]) / ([CyOGFP/+ ; UYi /+] + [CyOGFP/+ ; TM3/+])).
UYi lines located on the X chromosome were screened by crossing virgin [UYi/UYi ; +/+] or [UYi/FM0 ; +/+] female flies with [+/Y ; vg>bax/CyOGFP] males. If the mutant line was homozygous, the survival ratio was (([UYi/+ ; vg>bax/+]) / [UYi/+ ; CyOGFP/+]). If the mutant line was balanced with FM0, the ratio was (([UYi/+ ; vg>bax/+] + [FM0 /+ ; vg>bax/+]) / ([UYi/+ ; CyOGFP/+] + [FM0/+ ; CyOGFP/+]).
For UYi insertions located on autosomes, only crosses giving rise to a progeny of at least 25 individuals including at least two [vg>bax/UYi] were taken into account. For mutations of the X chromosome, only crosses giving rise to at least 25 individuals including at least 20 [UYi/+ ; CyOGFP /+] flies were taken into account.
To study phenotypes and survival, crosses were performed at 18°C and after five days they were either kept at 18°C or switched to 25°C. UYi mutations were considered as suppressors of lethality if the survival rate was higher than the observed average survival rate plus one standard deviation, either at 18°C or 25°C.. The average survival rate and standard deviation were independently determined for each of the three chromosomes bearing the UYi transgene to take into account the genetic background impact.
Classification of the wing phenotypes and the Wilcoxon test
All the lineages were analyzed in parallel with a control progeny and by a blind observer. Flies were classified according to their wing phenotype (strong, intermediate or weak) as previously described [26]. For flies showing two different wing phenotypes, the strongest phenotype of both wings was used for classification. In our screen, we have analyzed the distribution of graded phenotypes based on their expressivity. The Wilcoxon test was used to compare the distributions of the phenotypes between two lineages A and B [31]. The sign of the WsA‑B value determines whether the distribution A is stronger than B (Ws<0) or whether the distribution B is stronger than A (Ws>0). We considered the difference between A and B significant when αA‑B <10‑3.
Molecular characterization of UYi lines
To characterize the genes identified by screening for bax-induced phenotype modifiers, the DNA flanking the P[Mae-UAS.6.11] element was isolated by inverse polymerase chain reaction (PCR), essentially according to the Berkeley Drosophila Genome Project (BDGP) protocol (http://www.fruitfly.org/about/methods/inverse.pcr.html) and sequenced. The following primers were used in this study:
5’-GCAGTTGATTTACTTGGTTGCTGG-3’,5’-GGTAAGCTTCGGCTATCGAC-3’,5’-GCTTTCGCTTAGCGACGTGT-3’,5’-GCTTTCGCTTAGCGACGTG-3’,5’-GTATACTTCGGTAAGCTTCG-3’,5’-CTCTCAACAAGCAAACGTGC-3’,5’-ACACAACCTTTCCTCTCAACAA-3’,5’-GAATTGAATTGTCGCTCCGT-3’,5’-ATTGATTCACTTTAACTTGCAC-3’. Sequencing was performed by Genecust (Genopole, Evry, France). Sequences were submitted to BLAST search in the BDGP database to identify nearby genes.
Quantification of ptc-Gal4>(UAS-debcl)2-induced phenotype in the wing
To test the implication of Gpo-1 in debcl‑induced apoptosis, the severity of the wing tissue loss induced by UAS-debcl overexpression led by ptc-Gal4 driver was measured in different genetic backgrounds. We first verified that the Gpo1n322 LOF mutation did not induce any wing phenotype by itself. Then, ptc>debcl2, females were crossed with wild‑type males or males bearing a LOF mutation for Gpo-1. For each progeny, the distance between veins L3 and L4 was measured perpendicularly to the sixth sensilla of the dorsal row of the anterior wing margin and plotted against the distance between the extremity of veins 4 and 5. Student’s t-tests were then performed.
TUNEL staining
As previously described [60, 61], third instar larvae were dissected in PBS pH 7.6, fixed in PBS/formaldehyde 3.7%, washed three times for 10min in PBT (1X PBS, 0.5% Triton). Discs were then dissected and TUNEL staining was performed according to manufacturer’s instructions (ApopTag Red in situ apoptosis detection kit, Millipore, Temecula, CA, USA). Discs were mounted in CitifluorTM (Biovalley, Marne-La-Vallée, France) and observed with a Leica SPE upright confocal microscope (Leica, Wetzlar, Germany). White patches in the ptc expression domain were counted for at least 30 wing imaginal discs per genotype. Student’s t-tests were then performed.
Mitochondrial superoxide measurement
MitoSOX (Molecular Probes, Thermo Fisher Scientific) was used to measure the mitochondrial production of superoxide as described in [62]. Briefly, twenty wing imaginal discs were dissected in Schneider’s Drosophila medium (Fisher Bioblock scientific, Illkirch Graffenstaden, France), then cells were trypsinized. 5 μM MitoSOX were added to the cells placed at 37°C. Red fluorescence was then measured by flow cytometry in 2000 events per experimental condition. Flow cytometric measurement was performed using a BD LSRFortessa (Becton Dickinson, Franklin Lakes, USA). Fluorescence was induced by the Yellow‑Green Laser (561 nm). Red fluorescence was collected with a PE detector (emission: 578 nm).
ACKNOWLEDGEMENTS
This work was supported in part by grants from the Ligue Nationale Contre le Cancer (Comité des Yvelines). Jessie Colin, Cécile Milet and Amandine Clavier held a support from the Ministère de l’Enseignement Supérieur et de la Recherche. Jessie Colin, Julie Garibal, Cécile Milet and Amandine Clavier were supported by the Ecole Pratique des Hautes Etudes. We thank Laurent Théodore, Helena Richardson and the Bloomington Drosophila stock center for providing Drosophila strains. We thank François Juge and Martine Simonelig for precious information about the UY1131 strain and Magali Lecourtois for P[UY1615] localization.
![Table 2: Insertion site of the P[UAS] -element in suppressors of bax-induced phenotypes.](/article/68/asset/T2_thumb.png)

- 1. The Role of Mitochondria in Apoptosis. Annu Rev Genet. 2009; 43:95‑118. [PubMed] https://doi.org/10.1146/annurev-genet-102108-134850.
- 2. Bax activation and mitochondrial insertion during apoptosis. Apoptosis. 2007; 12(5):887‑896. [PubMed]
- 3. Structure of the CED‑4‑CED‑9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005; 437(7060):831‑837. [PubMed]
- 4. Regulation of apoptosis by C elegans CED‑9 in the absence of the C‑terminal transmembrane domain. Cell Death Differ. 2007; 14(11):1925‑1935. [PubMed] https://doi.org/10.1038/sj.cdd.4402215.
- 5. The mitochondrial TOM complex modulates bax‑induced apoptosis in Drosophila. Biochemical and biophysical research communications. 2009; 379(4):939‑943. [PubMed]
- 6. Inside an enigma: do mitochondria contribute to cell death in Drosophila? Apoptosis. 2009; 14(8):961‑968.. 2009; 379(4):939‑943. [PubMed] https://doi.org/10.1007/s10495-009-0362-6.
- 7. The mitochondrial pathways of apoptosis. Adv Exp Med Biol. 2012; 942:157‑183. [PubMed]
- 8. A cytochrome c-free fly apoptosome. Cell Death Differ. 2006; 13(7):1049‑1051. [PubMed]
- 9. Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila. Genes Dev. 2015; 29(3):277‑287. [PubMed] https://doi.org/10.1101/gad.255877.114.
- 10. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. Embo J. 2002; 21(13):3327‑3336. [PubMed] https://doi.org/10.1093/emboj/cdf354.
- 11. A GH3‑like domain in reaper is required for mitochondrial localization and induction of IAP degradation. J Biol Chem. 2003; 278(45):44758‑44768. . [PubMed]
- 12. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain‑lipid interactions. J Biol Chem. 2008; 283(1):367‑379. [PubMed]
- 13. Drosophila IAP antagonists form multimeric complexes to promote cell death. The Journal of cell biology. 2010; 190(6):1039‑1052. [PubMed] https://doi.org/10.1083/jcb.201004086.
- 14. Buffy, a Drosophila Bcl‑2 protein, has anti‑apoptotic and cell cycle inhibitory functions. EMBO J. 2003; 22(14):3568‑3579. [PubMed] https://doi.org/10.1093/emboj/cdg355.
- 15. Drosophila Bcl‑2 proteins participate in stress‑induced apoptosis, but are not required for normal development. Genesis. 2007; 45(4):184‑193. [PubMed]
- 16. Bax-like protein Drob‑1 protects neurons from expanded polyglutamine‑induced toxicity in Drosophila. Embo J. 2005; 24(14):2700‑2713. [PubMed] https://doi.org/10.1038/sj.emboj.7600721.
- 17. grim promotes programmed cell death of Drosophila microchaete glial cells. Mechanisms of development. 2010; 127(9‑12):407‑417. [PubMed] https://doi.org/10.1016/j.mod.2010.06.001.
- 18. Bcl‑2 proteins and autophagy regulate mitochondrial dynamics during programmed cell death in the Drosophila ovary. Development. 2011; 138(2):327‑338.
- 19. Alternative germ cell death pathway in Drosophila involves HtrA2/Omi, lysosomes, and a caspase‑9 counterpart. Developmental cell. 2013; 25(1):29‑42. [PubMed]
- 20. Debcl, a proapoptotic Bcl‑2 homologue, is a component of the Drosophila melanogaster cell death machinery. J Cell Biol. 2000; 148(4):703‑714. [PubMed]
- 21. Drob‑1, a Drosophila member of the bcl-2/CED-9 family that promotes cell death. Proc Natl Acad Sci USA. 2000; 97(2):662‑667. [PubMed] https://doi.org/10.1073/pnas.97.2.662.
- 22. The Drosophila bcl-2 family member dBorg‑1 functions in the apoptotic response to UV‑irradiation. Curr Biol. 2000; 10(9):547‑550. [PubMed]
- 23. Drosophila pro‑apoptotic Bcl‑2/Bax homologue reveals evolutionary conservation of cell death mechanisms. J Biol Chem. 2000; 275(35):27303‑27306. [PubMed]
- 24. The Bax/Bak ortholog in Drosophila, Debcl, exerts limited control over programmed cell death. Development. 2009; 136(2):275‑283. [PubMed] https://doi.org/10.1242/dev.019042.
- 25. Bcl‑2 and Bax mammalian regulators of apoptosis are functional in Drosophila. Cell Death and Differentiation. 2000; 7(9):804‑814.
- 26. reaper and bax initiate two different apoptotic pathways affecting mitochondria and antagonized by bcl‑2 in Drosophila. Oncogene. 2002; 21(42):6458‑6470. [PubMed]
- 27. Modulation of oxidative stress resistance in Drosophila melanogaster by gene overexpression. Genesis. 2002; 34(1‑2):76‑79. [PubMed]
- 28. Gain-of-function screen identifies a role of the Src64 oncogene in Drosophila mushroom body development. J Neurobiol. 2003; 57(3):291‑302. [PubMed]
- 29. A new genetic locus controlling growth and proliferation in Drosophila melanogaster. Genetics. 2003; 164(3):1015‑1025. [PubMed]
- 30. The HIV-1 Vpu protein induces apoptosis in Drosophila via activation of JNK signaling. PLoS One. 2012; 7(3):e34310. [PubMed] https://doi.org/10.1371/journal.pone.0034310.
- 31. Individual comparisons by ranking methods. Biometrics. 1945; 1:80‑83. [PubMed]
- 32. The genome sequence of Drosophila melanogaster. Science. 2000; 287(5461):2185‑2195. [PubMed]
- 33. The drosophila Bcl‑2 family protein Debcl is targeted to the proteasome by the beta‑TrCP homologue slimb. Apoptosis. 2014; 19(10):1444‑1456. [PubMed]
- 34. Modification of glycolysis affects cell sensitivity to apoptosis induced by oxidative stress and mediated by mitochondria. Biochem Biophys Res Commun. 2004; 313(4):984‑991. [PubMed]
- 35. Mitochondria and reactive oxygen species. Which role in physiology and pathology? Adv Exp Med Biol. 2012; 942:93‑136. [PubMed]
- 36. Superoxide and hydrogen peroxide production by Drosophila mitochondria. Free Radic Biol Med. 2003; 35(8):938‑948. [PubMed]
- 37. Genome-wide screen for modifiers of ataxin‑3 neurodegeneration in Drosophila. PLoS Genet. 2007; 3(10):1950‑1964. [PubMed] https://doi.org/10.1371/journal.pgen.0030177.
- 38. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001; 15(6):762‑773. [PubMed] https://doi.org/10.1101/gad.870801.
- 39. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006; 133(14):2639‑2648. [PubMed]
- 40. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self‑renewal. Dev Cell. 2006; 10(4):441‑449. [PubMed]
- 41. Mei‑P26 Mediates Tissue-Specific Responses to the Brat Tumor Suppressor and the dMyc Proto‑Oncogene in Drosophila. Genetics. 2014.
- 42. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol Cell. 1999; 4(3):309‑319. [PubMed]
- 43. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation‑induced apoptosis in vivo. Cancer Cell. 2002; 2(4):279‑288. [PubMed]
- 44. Slug antagonizes p53‑mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005; 123(4):641‑653. [PubMed]
- 45. Repression of Puma by Scratch2 is required for neuronal survival during embryonic development. Cell death and differentiation. 2011. [PubMed] https://doi.org/10.1038/cdd.2010.190.
- 46. The axon guidance gene lola is required for programmed cell death in the Drosophila ovary. Dev Biol. 2007; 304(2):771‑785. [PubMed] https://doi.org/10.1016/j.ydbio.2007.01.029.
- 47. Inhibitor of apoptosis 2 and TAK1‑binding protein are components of the Drosophila Imd pathway. Embo J. 2005; 24(19):3423‑3434. [PubMed] https://doi.org/10.1038/sj.emboj.7600807.
- 48. Lola regulates midline crossing of CNS axons in Drosophila. Development. 2002; 129(6):1317‑1325. [PubMed]
- 49. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006; 439(7075):430-436. [PubMed]
- 50. Links between metabolism and apoptosis in mammalian cells: applications for anti‑apoptosis engineering. Metab Eng. 2007; 9(4):317‑326. [PubMed]
- 51. Involvement of cytochrome c oxidase subunits Va and Vb in the regulation of cancer cell metabolism by Bcl‑2. Cell Death Differ. 2009:doi: 10.1038/cdd.2009.1132. [PubMed] https://doi.org/10.1038/cdd.2009.1132.
- 52. Mitochondrial reactive oxygen species in cell death signaling. Biochimie. 2002; 84(2‑3):131‑141. [PubMed]
- 53. Utilization of yeast to investigate the role of lipid oxidation in cell death. Antioxid Redox Signal. 2004; 6(2):259‑267. [PubMed]
- 54. Role of oxidative phosphorylation in Bax toxicity. Mol Cell Biol. 2000; 20(10):3590‑3596. [PubMed]
- 55. Bcl‑x(L) complements Saccharomyces cerevisiae genes that facilitate the switch from glycolytic to oxidative metabolism. J Biol Chem. 2002; 277(47):44870‑44876.
- 56. Generation of hydrogen peroxide by brown adipose tissue mitochondria. J Bioenerg Biomembr. 1987; 19(4):397‑407. [PubMed]
- 57. Mitochondrial FAD‑linked Glycerol‑3‑phosphate Dehydrogenase: A Target for Cancer Therapeutics. Pharmaceuticals (Basel). 2014; 7(2):192‑206. [PubMed] https://doi.org/10.3390/ph7020192.
- 58. A stable genomic source of P element transposase in Drosophila. Genetics. 1988; 118(3):461‑470. [PubMed]
- 59. New mutations in the Wilson disease gene, ATP7B: implications for molecular testing. Genet Test. 2008; 12(1):139‑145. [PubMed]
- 60. Mutating RBF can enhance its pro‑apoptotic activity and uncovers a new role in tissue homeostasis. PLoS One. 2014; 9(8):e102902. [PubMed] https://doi.org/10.1371/journal.pone.0102902.
- 61. The pro‑apoptotic activity of Drosophila Rbf1 involves dE2F2‑dependent downregulation of diap1 and buffy mRNA. Cell Death Dis. 2014; 5:e1405. [PubMed] https://doi.org/10.1038/cddis.2014.372.
- 62. Mitochondrial p53 mediates a transcription‑independent regulation of cell respiration and interacts with the mitochondrial F1F0‑ATP synthase. Cell Cycle. 2013; 12(17):2781‑2793. [PubMed] https://doi.org/10.4161/cc.25870.
Last Modified: 2016-06-03 21:38:44 EDT
PII: 68
