Genes & Cancer
A novel DNA damage response mediated by DNA mismatch repair in Caenorhabditis elegans: induction of programmed autophagic cell death in non-dividing cells
Takahito Moriwaki1, Yuichi Kato1, Chihiro Nakamura1, Satoru Ishikawa1 and Qiu-Mei Zhang-Akiyama1
1 Laboratory of Stress Response Biology, Graduate School of Science, Kyoto University, Kitashirakawa-Oiwakecho, Sakyo-ku, Kyoto, Japan
Correspondence to: Qiu-Mei Zhang-Akiyama, email: [email protected]
Keywords: MLH1, ATM, ATR, Organism death, Cancer
Received: December 17, 2014
Accepted: June 28, 2015
Published: June 29, 2015
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
DNA mismatch repair (MMR) contributes to genome integrity by correcting errors of DNA polymerase and inducing cell death in response to DNA damage. Dysfunction of MMR results in increased mutation frequency and cancer risk. Clinical researches revealed that MMR abnormalities induce cancers of non-dividing tissues, such as kidney and liver. However, how MMR suppresses cancer in non-dividing tissues is not understood.
To address that mechanism, we analyzed the roles of MMR in non-dividing cells using Caenorhabditis elegans (C. elegans), in which all somatic cells are non-dividing in the adult stage. In this study, we used stable MMR-mutant lines with a balancer chromosome. First, we confirmed that deficiency of MMR leads to resistance to various mutagens in C. elegans dividing cells. Next, we performed drug resistance assays, and found that MMR-deficient adult worms were resistant to SN1-type alkylating and oxidizing agents. In addition, dead cell staining and reporter assays of an autophagy-related gene demonstrated that the cell death was autophagic cell death. Interestingly, this autophagic cell death was not suppressed by caffeine, implying that MMR induces death of non-dividing cells in an atl-1-independent manner. Hence, we propose the hypothesis that MMR prevents cancers in non-dividing tissues by directly inducing cell death.
INTRODUCTION
DNA mismatch repair (MMR) ensures genome integrity by correcting DNA replication and recombination errors [1]. MMR is highly conserved from bacteria to humans [2, 3]. In eukaryotes, MMR starts with detection of mismatches by MSH2-MSH6 heterodimer (MutSα) followed by interacting with MLH1-PMS2 heterodimer (MutLα), proliferating cell nuclear antigen (PCNA), and exonuclease1 (EXO1), and then that complex excises the newly synthesized strand by exonuclease activity of EXO1[1]. Dysfunction in MMR results in an increased mutation frequency in many kinds of organisms [4]. MMR abnormalities in humans result in increased risks of cancer, including hereditary nonpolyposis colorectal cancer (HNPCC) [5]. It is considered that MMR-deficient colorectal cells rapidly accumulate mutations due to their high turnover, which ultimately induces carcinogenesis. On the other hand, it has been reported that dysfunction of MMR also leads to increased cancer risks in non-dividing tissues, such as kidney and liver [6, 7]. However, it is not understood how MMR suppresses carcinogenesis in non-dividing cells.
Recent studies have revealed that MMR plays key roles in the DNA damage response (DDR) process [8]. An in vitro analysis revealed that MutSα has affinity to O6-methylguanine, which is generated by SN1-type DNA alkylation [9]. Furthermore, an in vivo analysis using mammalian cultured cells showed that following recognition of the DNA lesions, MMR activates the G2/M checkpoint or apoptosis pathway via the ataxia telangiectasia and Rad3-related protein (ATR)/ATR Interacting Protein (ATRIP) pathway after forming a complex with MutLα, PCNA, and EXO1 [10–12]. Due to the lack of that apoptotic pathway, MMR-deficient cells show resistance to SN1-type alkylating agents [13]. In addition to O6-methylguanine, a wide range of DNA lesions have been identified as substrates of MMR, such as apurinic/apyrimidinic (AP) sites, interstrand cross-links, 8-oxoguanine, and UV-photoproducts [14-17]. Although some reports have shown the possibility that MMR can also function in non-dividing cells, for example, by noncanonical MMR, the physiological function thereof is not well understood [18, 19].
In order to address the relationship between the dysfunction of MMR and carcinogenesis in non-dividing cells, we investigated the roles of MMR in non-dividing cells using the nematode Caenorhabditis elegans (C. elegans). C. elegans has been extensively used as a model organism for aging, the nervous system, and apoptosis. The cell fates of every cell in C. elegans have been completely determined, and all the somatic cells of an adult worm are non-dividing [20, 21]. Also in C. elegans, dysfunction of the MMR gene results in increased mutation frequency; for instance, knockdown of mlh-1, the homologue of hMlh1, results in increased mutation frequency [22].
Recently, it was reported that in response to 5-fluorouracil (5-FU) treatment, MMR induces expression of lgg-1 (C. elegans MAP1LC3 orthologue), one of the autophagosome genes in a manner dependent on ATL-1 (C. elegans ATR homologue) and two AP-endonucleases, EXO-3 and APN-1 in C. elegans embryos [23]. 5-FU treatment inhibits thymidine synthetase and incorporation of 5-fluorodeoxyuridine in genomic DNA [24]. In that pathway, MMR induces autophagic cell death in response to 5-FU treatment in embryos, indicating that C. elegans MMR recognizes 5-fluorodeoxyuridine in genomic DNA and induces autophagic cell death. However, other possible substrates that could similarly induce autophagy have not been identified yet in C. elegans. In the present study, we performed drug treatments of N2 and mlh-1(ok1917) mutant worms at various developmental stages. mlh-1(ok1917) worms have been used previously for analysis of the relationship between MMR and other DNA repair pathways [25, 26]. We first performed screenings of substrates of C. elegans MMR using pachytene stage meiotic cells, in which apoptosis is induced in response to DNA damage [27]. The results revealed that mlh-1(ok1917) pachytene stage meiotic cells showed decreased apoptotic cells in response to various DNA lesions, like mammalian cultured cells. In addition, we confirmed by L1 drug resistance assays that MMR induces cell cycle arrest or cell death in response to DNA damage in dividing somatic cells. We next performed drug resistance assays using adult worms, and found that mlh-1(ok1917) adult worms showed resistance to alkylating and oxidizing agents. In addition, the results of reporter assays of lgg-1, one of the autophagosome genes, suggested that the cell death of non-dividing adult somatic cells was not apoptosis but autophagic cell death. Interestingly, this autophagic cell death was not suppressed by caffeine treatment, suggesting that autophagic cell death is independent on ATL-1. These results clearly demonstrate that C. elegans MMR can induce cell death in non-dividing cells in a manner apparently different from that in embryos, and suggest that MMR prevents carcinogenesis in non-dividing tissues by inducing cell death.
RESULTS
Establishment of a stable MMR mutant worm line
Previous studies showed that dysfunction of MMR results in increased mutation frequency in C. elegans [22]. Backcrossed msh-2(ev679::Tc1) worms have no abnormalities in basic phenotypes, such as growth and lifespan. However, they die out during serial passage in culture, because of accumulation of mutations [28]. These facts mean that a stable maintenance system for MMR mutants is required to analyze the function of MMR in DDR without disturbing background mutations. In previous studies, it was shown that an increased mutation frequency is not observed in MMR heterozygous mutants in various species [29, 30]. Therefore, we first established a stable maintenance system for the MMR heterozygous mutant using a balancer chromosome, hT2 (Supplementary Figure S1). hT2 is one of the balancer chromosomes that prevents parts of chromosomes I and III from homologous recombination. In addition, the myo-2p::GFP gene and recessive lethal gene were inserted as markers, and we could maintain the heterozygous mutants by selective picking of GFP-positive worms [31].
C. elegans MLH-1 is a component of C. elegans MutLα, and knockdown of C. elegans mlh-1 results in increased mutation frequency [22]. We performed backcrossing of the mlh-1-deficient strain RB1572 (mlh-1(ok1917)), with N2 worms (wild-type). After backcrossing, we crossed the worms with JK2739 (lin-6(e1466) dpy-5(e61) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III)) worms to maintain the MMR mutant line in genetically stable condition (mlh-1(ok1917/hT2)). We maintained mlh-1(ok1917/hT2) worms by picking GFP-positive worms until use and isolated mlh-1(ok1917/ok1917) worms for experiments by picking GFP-negative worms (Supplementary Figure S1). We confirmed the absence of abnormalities in basic phenotypes in the backcrossed mlh-1(ok1917) mutant line (Supplementary Figure S2).
mlh-1(ok1917) pachytene stage meiotic cells showed decreased apoptosis in response to a wide range of DNA-damaging agents
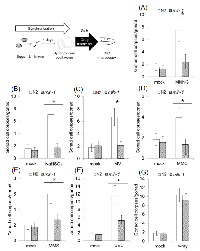
In previous research, it was reported that MMR induces autophagic cell death in response to 5-FU treatment in a manner dependent on two AP endonucleases, EXO-3 and APN1, in C. elegans embryos [23]. 5-FU induces genome instability by incorporation of 5-fluorodeoxyuridine into genomic DNA during DNA replication [32]. Therefore, 5-FU is not suitable for our research focused on the function of MMR in the DNA damage response in non-dividing cells. In order to identify other possible substrates, we measured the number of apoptotic pachytene stage meiotic cells. In C. elegans, apoptosis in pachytene stage meiotic cells is frequently induced by DNA damage and causes the formation of condensed structures in the gonad that are easily detected by DIC microscopy [33]. We treated synchronized adult worms for 24 hours with various drugs, and measured the number of apoptotic cell corpses of pachytene stage meiotic cells.
N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) is an SN1-type alkylating agent that is frequently used in research on MMR-dependent cell death and cell cycle arrest. MNNG mainly generates O6-methylguanine via SN1-type alkylation in genomic DNA [9]. MNNG treatment increased the number of apoptotic pachytene stage meiotic cells in N2 gonad arms, but such an increase was not observed in the mlh-1(ok1917) gonad arms (Figure 1A). These results indicate that C. elegans MMR induces apoptosis in response to MNNG treatment, as it does in mammalian cultured cells.
In addition, we also tested the following types of DNA-damaging agents: SN2-type alkylating agent, deaminating agent, oxidizing agent, cross-linking agent, UVC, and γ-rays. Methyl methanesulfonate (MMS) is an SN2-type alkylating agent, and in contrast to SN1-type alkylating agents mainly generates unstable DNA lesions such as 7-methylguanine and 3-methyladenine, which are easily converted to AP sites [34]. Mitomycin C (MMC) is a cross-linking agent that generates interstrand cross linking in genomic DNA [35]. Methyl viologen (MV), which is also called paraquat, is a superoxide-inducing agent, and generates oxidative DNA lesions in genomic DNA [36, 37]. Sodium hydrogen sulfite (NaHSO3) is a deaminating agent that generates deaminated DNA lesions such as uracil and 5-(hydroxymethyl) uracil in genomic DNA [38]. UVC irradiation induces dimerization of pyrimidine bases [39]. These DNA-damaging agents have been reported to be activators of MMR in previous studies [9, 14-17]. Pachytene stage meiotic cells of mlh-1(ok1917) showed less apoptosis than those of N2 worms in response to all of the types of DNA-damaging treatments which we tested, except γ-rays irradiation (Figures 1B-F). These results suggest that C. elegans MMR can induce cell death in response to various base modifications. However, γ-rays irradiation induced a similar increase of death of pachytene stage meiotic cells in N2 as in mlh-1(ok1917) (Figure 1G). This is considered to have been due to the cytotoxicity of DNA double-strand breaks (DSBs) induced by γ-rays.
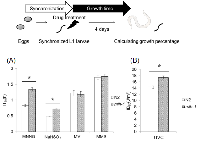
C. elegans MMR regulated larval development
Microarray experiments revealed that C. elegans MMR genes msh-2, msh-6, mlh-1 and pms-2 are abundantly expressed in pachytene stage meiotic cells and embryos, and their expression levels are decreased after hatching by approximately 10-fold, except for that of pms-2 [40, 41]. Next, we examined whether C. elegans MMR induces cell death or cell cycle arrest in post-embryonic cells, dividing somatic cells (dSCs), using L1 larvae. C. elegans larvae develop to adult worms through 4 stages or molts, which are designated as L1, L2, L3 and L4. We synchronized worms at L1 stage and performed a time-course of treatment with various DNA-damaging agents. Four days after the drug treatments, we measured the growth (percentage) from L1 larvae to the L4-adult stages, and calculated the half maximal inhibitory time (IT50) or dose (ID50) from three-parameter logistic curves (Supplementary Figure S3). mlh-1(ok1917) larvae showed significantly higher IT50 or ID50 in response to MNNG, NaHSO3, and UVC than N2 larvae (Figures 2A and B). This suggests that C. elegans MMR induces cell cycle arrest or cell death in dSCs. In contrast, no significant difference was observed in the sensitivity to MV or MMS between N2 and mlh-1(ok1917) (Figure 2A). Such a difference was observed for the msh-6(pk2504) mutant (Supplementary Figures S4A and B). The observed difference of response suggests that there may be differential responses to DNA damage between pachytene stage meiotic cells and proliferating larval cells, although this difference may just be due to differences of drug penetrance or replication stress. In addition, we tested MMC and γ-rays, and found that they did not affect percentage growth significantly at the maximum doses used, 0.2 mg/ml and 100 Gy, respectively, in either N2 or mlh-1(ok1917) L1 larvae (data not shown).
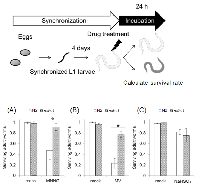
C. elegans MMR induced non-dividing cell death in response to DNA damage
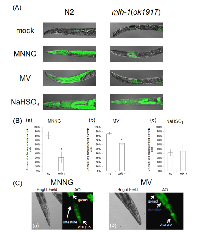
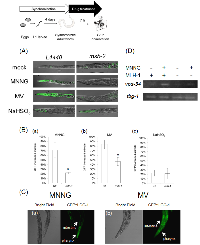
We confirmed that C. elegans MMR induces cell death or cell cycle arrest in post embryonic cells, dSCs, in response to some mutagens, including MNNG. Next, we examined the drug resistance of non-dividing somatic cells (ndSCs) using adult worms. We prepared fully matured adult worms by culturing synchronized L1 larvae on NGM plates for 4 days, and treated them with DNA-damaging agents for 1 hour. 24 hours after the drug treatments, we measured survival rate. As shown in Figures 3A and B, mlh-1(ok1917) adult worms showed a significantly higher survival rate than N2 worms after the MNNG and MV treatments. Moreover, no significant difference of the sensitivity to NaHSO3 treatment was observed (Figure 3C).In order to address whether C. elegans MMR induces ndSCs’ death, we performed acridine orange (AO) staining. AO is a non-fluorescent dye that shows fluorescence when it binds stably to nucleic acids. Because AO is actively exported out of living cells, fluorescence is observed only in dead cells after sufficient destaining [42]. As shown in Figures 4A and B, there were significantly fewer worms with AO fluorescence in mlh-1(ok1917) than in N2 in the cases of MNNG and MV treatment. In contrast, the same levels of AO fluorescence were observed in the case of NaHSO3 treatment (Figures 4A and B). Moreover, AO-induced fluorescence was especially observed in the pharynx and intestines of N2 worms after MV treatment, whereas there was no such localization of the fluorescence of mlh-1(ok1917) worms (Figure 4A). As shown in Figure 4C, MNNG treatment induced not only pharynx cell death but also death of the surrounding cells, while MV treatment induced only pharynx cell death in the head part. In C. elegans, the pharynx is in the head part, and neurons are located surrounding the pharynx. These results suggest that MNNG and MV especially injure neurons and intestine, respectively, in an MMR-dependent manner in C. elegans.
C. elegans MMR induced autophagic cell death in ndSCs independently of ATL-1
In humans, MMR induces apoptosis or cell cycle arrest via the ATR/ATRIP pathway [11]. In response to damage recognition by MutSα, ATR/ATRIP is activated and phosphorylates CHK1 to induce cell cycle arrest and CHK2 to induce apoptosis [11, 12]. In C. elegans, this system is well conserved, and it has been reported that in C. elegans embryos, MMR activates CHK-1 (C. elegans CHK1 homologue) and induces autophagic cell death in response to 5-FU treatment in an ATL-1-dependent manner [23]. In order to examine how MMR induces cell death in ndSCs, we next tried to identify the type of cell death. In C. elegans, the cascade of apoptosis is well understood, and is regulated by only 4 factors: CED-3, CED-4, CED-9, and EGL-1. CED-3 is the effector caspase, and is converted to the active form by CED-4. Usually, CED-4 is blocked by CED-9, but only when cells undergo apoptosis, CED-4 activates CED-3 as a result of inhibition of CED-9 by EGL-1. egl-1 is one of the genes whose expression is regulated by CEP-1 (the C. elegans p53 homologue) and is responsible for apoptosis in pachytene stage meiotic cells [43]. To determine whether the cell deaths in ndSCs were due to apoptosis, we measured somatic egl-1 mRNA levels in MNNG-treated worms by semi-quantitative RT-PCR using the temperature-sensitive sterile mutant glp-4(bn2) cultured at 25° C, which does not have gonads, but no significant induction of egl-1 mRNA was detected, whereas Wt worms, which have gonads, showed increased egl-1 mRNA in response to MNNG treatment (Supplementary Figure S5).
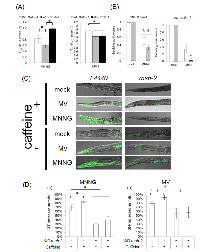
We then focused on autophagic cell death. Autophagic cell death is also known as type 2 cell death, and is defined as cell death with autophagy rather than apoptosis [44, 45]. In humans, MAP1LC3 (ATG8) is widely used as a marker of autophagic cell death. In mammalian cells, MAP1LC3 exists in two forms: full-length form MAP1LC3-I (MAP1LC3) and shorter-form MAP1LC3-II [45]. During autophagy, MAP1LC3-I is transformed to MAP1LC3-II by cleavage, and MAP1LC3-II localizes to membranes of autophagosomes [45]. In the process of autophagic cell death, MAP1LC3 is specifically upregulated [44, 45]. This system is well conserved also in C. elegans, and upregulation of lgg-1 in the process of autophagic cell death has been reported [23]. Based on these facts, we used lgg-1 expression as a marker of autophagic cell death, and we performed reporter assays using MAH236 (lgg-1p::GFP::lgg-1) worms. Because of the expressed GFP::LGG-1, we could not use a stable maintenance system using a GFP balancer. Therefore, we performed reporter assays using msh-2 knockdown worms (Supplementary Figure S6). As shown in Figures 5A and B, the expression level of lgg-1 was elevated by MNNG and MV treatments, and knockdown of msh-2 suppressed this induction, whereas there was no difference between the NaHSO3-treated control and msh-2 knockdown worms. LGG-1 was expressed in the pharynx and intestine in MV-treated N2 worms, but only in the pharynx and surrounding cells in MNNG-treated N2 worms (Figure 5C). In addition, we analyzed the mRNA levels of vps-34, a homologue of human class III PI3K, which is expressed during autophagic cell death in C. elegans, and we confirmed that mRNA levels of vps-34 were increased in response to MNNG treatment (Figure 5D) [23]. These results suggest that MMR induces autophagic cell death in ndSCs in response to MNNG and MV treatment.
There are two hypotheses about how MMR induces cell death: “futile repair” and “signaling” [8]. In futile repair, repeated excision and resynthesis occur, and they lead to DSBs and genome instability [46]. It has been reported that MNNG generates DSBs in an MMR-dependent manner, and a DSB repair mutant shows sensitivity to MNNG [47, 48]. Ataxia telangiectasia mutated (ATM) is a member of the PIKK family, and it is required for DSB repair, homologous recombination, non-homologous end-joining, and alternative end-joining [49]. In C. elegans, ATM-1, a homologue of ATM, also plays a critical role in DSB repair [50, 51]. In order to analyze whether DSBs are generated by MNNG treatment, we performed drug resistance assays using atm-1(tm5027) worms. In C. elegans, atm-1(tm5027) L1 larvae showed sensitivity to both MNNG and MMS, but the sensitivity to MNNG was abrogated by deficiency of mlh-1, whereas the sensitivity to MMS was not (Supplementary Figure S7, Figure 6A). MNNG generates relatively stable DNA lesions such as O6-methylguanine, whereas MMS induces DSBs via accumulation of AP sites, which are converted to SSBs by AP endonucleases. From these facts, it is considered likely that MNNG induced DSBs in an MMR-dependent manner in dSCs, suggesting the presence of futile repair in dSCs. In contrast, adult worms did not show sensitivity to MNNG although they showed sensitivity to MMS (Figure 6B). This means that MNNG can not induce DSB in ndSCs, suggesting the absence of futile repair in ndSCs. Therefore, C. elegans MMR can induce cell death via both futile repair and signaling depending on the types of cell or stages.
During futile repair, in order to protect the template single-strand DNA, replication protein A (RPA) is accumulated on the template strand. This RPA recruits ATR, and the recruited ATR activates the signaling cascade [52]. This suggests that ATR may not be required for “signaling” cell death. Accordingly, we examined whether ATL-1 is responsible for ndSCs death. In C. elegans, the knockout of atl-1 results in lethality. For this reason, we can not use an atl-1 null mutant, although we can use an atl-1 heterozygous mutant (tm853/nT1). This means that we could only decrease the atl-1 expression level to only about 50% by either knockout or knockdown. In order to more efficiently inhibit ATL-1, we performed caffeine treatment. Caffeine is widely used as an inhibitor of ATM and ATR proteins, and in a previous study, treatment with 20 mM caffeine for at least 6 hours decreased the ATL-1 activity in C. elegans [53]. In our experiments, caffeine treatment did not inhibit expression of LGG-1, but rather appeared to slightly promote it under conditions of MNNG treatment (Figure 6C and D). These results suggest that ATL-1 is not responsible for autophagic cell death in ndSCs.
DISCUSSION
In the present study, we demonstrated that C. elegans MMR induces autophagic cell death in response to DNA damage in non-dividing cells. Previous studies showed that MMR-induced cell death and cell cycle arrest are responsible for genome integrity and cancer prevention in mammalian dividing cells [4]. On the other hand, some reports showed that MMR factors are also present in non-dividing cells and MMR prevents carcinogenesis in non-dividing cells, such as kidney and liver [54]. Some reports showed DNA replication-independent functions of MMR, such as non-canonical MMR, which usually result in inaccurate DNA repair and induce mutations and cancers [18, 19]. These facts are not in accord with clinical observations, and how MMR prevents cancers of non-dividing cells has remained unclear. Our finding that MMR induces cell death in non-dividing cells in response to DNA damage suggests one possible function of MMR in the prevention of carcinogenesis in non-dividing cells.
A recent study showed that 5-FU treatment induces autophagic cell death in an MMR- and AP endonuclease-dependent manner in C. elegans embryos, but other substrates of C. elegans MMR have not been identified [23]. In order to analyze the functions of C. elegans MMR, we first tried screening the substrates of C. elegans MMR using pachytene stage meiotic cells. These screenings showed that mlh-1(ok1917) pachytene stage meiotic cells have a deficiency of apoptosis induced by a wide range of DNA-damaging agents, such as MNNG and UVC (Figure 1). This suggests that C. elegans MMR, similarly to MMR in mammalian cultured cells, induces apoptosis in response to a wide range of DNA lesions, for example, O6-methylguanine, AP sites, and thymine dimers [9,14–17]. In contrast, mlh-1(ok1917) pachytene stage meiotic cells showed induction of normal levels of apoptosis by γ-rays irradiation (Figure 1G). While γ-rays irradiation generates reactive oxygen species (ROS), such as superoxide, γ-rays irradiation also directly induces DNA double-strand breaks [55]. As shown in Figure 1C, mlh-1(ok1917) pachytene stage meiotic cells can undergo induction of apoptosis in response to MV, which generates superoxide, indicating that C. elegans MMR responds to oxidative DNA damage caused by superoxide. Thus, γ-rays irradiation induced cell death not in response to oxidative lesions, but rather in response to more toxic DSBs.
In C. elegans, MMR factors are abundantly expressed in germ cells and embryos, while they are not as highly expressed in postembryonic somatic cells [40, 41]. To examine whether C. elegans MMR works in postembryonic somatic cells, we measured the drug resistance during growth, and found that the development of mlh-1(ok1917) larvae showed higher IT50 or ID50 in response to some kinds of DNA-damaging agents compared to N2 larvae (Figures 2A and B). This means that C. elegans MMR induces cell death or cell cycle arrest in dividing somatic cells (dSCs), and deficiency of MMR results in DNA-damage-resistant growth of L1 larvae.
Next, we performed drug resistance assays using adult worms, and found that mlh-1(ok1917) adult worms are resistant to MNNG and MV treatments (Figures 3A and B). This suggests that C. elegans MMR induces non-dividing somatic cells’ (ndSCs’) death. In order to confirm whether MMR induces ndSCs’ death, we performed AO staining. The results of AO staining suggested that C. elegans MMR induced cell death in response to the drug treatments in ndSCs such as those in pharynx, intestine and neurons (Figure 4). These results indicate that C. elegans MMR induces cell death independently of DNA replication.
In addition, we obtained interesting findings about the mechanism of cell death induced by MMR. In eukaryotic organisms, two models have been suggested for the mechanisms underlying MMR-induced cell death: “futile repair” and “signaling” [8]. In a previous study, it was shown that EXO1 is essential for inducing cell death by MMR in mammalian cultured cells, but the exonuclease activity of EXO1 is not required [10]. This suggests that MMR can induce cell death just by signaling, without futile repair. In our experiments, atm-1(tm5027) L1 larvae showed sensitivity to both MNNG and MMS (Figure 6A). On the other hand, atm-1(tm5027) adult worms did not show sensitivity to MNNG, whereas they were sensitive to MMS (Figure 6B). These results suggest that MNNG treatment generates DSBs in dSCs, but not in ndSCs. In addition, the sensitivity to MNNG in L1 larvae was abrogated by deficiency of MLH-1 (Figure 6A). Based on these facts, we hypothesize that MMR induces cell death by futile repair and/or signaling in dividing cells, and by only signaling in non-dividing cells.
In futile repair, RPA is accumulated on single-strand DNA, and RPA recruits ATR/ATRIP. Therefore, the fact that MMR can induce cell death by just signaling suggests that MMR can induce cell death independently of ATR/ATRIP in non-dividing cells. To examine the necessity for ATR/ATRIP in ndSCs’ death, we performed caffeine-inhibition assays of the induction of LGG-1 using adult worms. The results of caffeine inhibition assays suggested that C. elegans MMR induced autophagic cell death independently of ATL-1, which means that C. elegans MMR induced autophagic cell death in ndSCs in an apparently different manner from that in embryos (Figures 6C and D) [23].
In HeLa cells, the MAPO1/AMPK pathway has been identified as a pathway that induces apoptosis in response to SN1-type alkylating agents in an MMR-dependent manner [56]. These factors are highly conserved among higher eukaryotes, and also in C. elegans. The fact that p53 is inactivated in HeLa cells implies that the MAPO1/AMPK pathway may also not require ATR/ATRIP. Furthermore, AMPK regulates autophagy via inhibition of mTOR, whose activity is not inhibited by caffeine, and indeed rather is promoted by it [57]. This is in accord with the results of our caffeine inhibition assay (Figure 6D), and implies that C. elegans MMR induces autophagic cell death via AMPK/mTOR in ndSCs.
As another possibly significant feature of autophagy induced by MMR, it is considered that MMR may prevent the accumulation of abnormal proteins. DNA lesions can induce not only replication errors but also transcription errors. Therefore, severe damage of genomic DNA leads to accumulation of abnormal proteins. MMR may contribute to the maintenance of cell function by surveying DNA lesions and inducing autophagy to degrade abnormal proteins.
We obtained the interesting findings that the sensitivity of mlh-1(ok1917) cells differed among pachytene stage meiotic cells, dSCs, and ndSCs (Figures 1-3). In addition, MMR induced autophagic cell death in different tissues in response to MNNG versus MV (Figures 4 and 5). In humans, MutSα has SQ/TQ sites, and the kinase inhibitor UCN-01 decreases the phosphorylation levels of MutSα and its binding affinity to O6-methylguanine [9]. Even though our findings might have been due simply to differences of drug penetration or just the difference of the presence or absence of DNA replication, which induces genome instability via replication stress, these differences may mean that MMR components may be appropriately modified depending on the growth stage or tissue in order to efficiently and appropriately protect each tissue. For example, C. elegans MMR can recognize more kinds of DNA damage in pachytene stage meiotic cells than in somatic cells, and this may be due to the various modifications of C. elegans MutSα in order to enable precise inheritance of the genomic information.
Recently much attention has been paid to dormant cancer cells. These cells are considered to be cancer stem cells and to show resistance to many anticancer agents. These cells are thought to be a cause of recurrent carcinoma because of the difficulty associated with killing them [58]. This resistance may be derived from differences in the DDR pathways between dividing and non-dividing cells. DDR pathways in non-dividing cells need to be examined in more detail. The results of the present study may represent a breakthrough for cancer therapy for dormant cancer stem cells.
MATERIALS & METHODS
Construction of a knockdown vector
The sequence information of H26D21.2 (Cemsh-2) was obtained from the NCBI database. PCR was conducted using two PCR primers listed in the Supplementary Table and KOD-Plus- polymerase (TOYOBO, Japan). The amplified products were digested with Bgl II and Hind III for cloning an approximately 1-Kbp fragment from the 5’ end. The digested products were then ligated to the L4440 vector digested with BamH I and Hind III.
C. elegans strains and culture conditions
The wild-type strain (Bristol N2), RB1572[mlh-1(ok1917) III], JK2739[lin-6(e1466) dpy-5(e61) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III)], NL2511[msh-6(pk2504) I], MAH236; [lgg-1p::GFP::lgg-1 + odr-1p::RFP]), and SS104 [glp-4(bn2)I] were supplied by the Caenorhabditis Genetics Center (Minneapolis, USA). atm-1(tm5027) mutant was supplied by the National BioResource Project (Tokyo, Japan). A deletion was verified in the mlh-1 and atm-1 genes by PCR using two primer pairs listed in the Supplementary Table. RB1572, NL2511 and atm-1(tm5027) mutant worms were backcrossed with Bristol N2 two times and maintained with the GFP balancer hT2 to avoid the accumulation of mutations [31]. lgg-1 reporter strain (tm5027 [lgg-1p::GFP::lgg-1 + odr-1p::RFP]), atm-1(tm5027)/mlh-1(ok1917) double mutant and balanced germline mutant [mlh-1(ok1917) glp-4(bn2) lin-6(e1466) dpy-5(e61) I/hT2 [bli-4(e937) let-?(q782) qIs48] ], were generated by crossing each strain. Worms were cultured at 20°C on 50-mm NGM plates containing 0.3% (w/v) NaCl, 0.25% (w/v) polypeptone, 0.005% (w/v) cholesterol, 1 mM MgSO4, 1 mM CaCl2, 25 mM potassium phosphate (pH 6.0), and 0.17% (w/v) agar with a lawn of Escherichia coli (E. coli) OP50 [59].
Synchronization of worms
Starved L1 larvae were prepared in order to obtain synchronized worms. Briefly, worms on NGM plates were harvested by washing out with distilled water and incubated in alkaline hypochlorite [500 mM NaOH and 1.2% (v/v) hypochlorite] until their bodies were completely dissolved (5-10 minutes). Eggs were then washed three times with S basal [50 mM potassium phosphate, (pH 6.0), containing 100 mM NaCl]. The eggs were hatched and synchronized by incubation at 20° C overnight without food [59].
Measurement of lifespan
Synchronized L1 larvae were cultured on NGM plates until they developed to the adult stage (3 days). To determine lifespans, worms were harvested with M9 buffer (22 mM KH2 PO4, 42 mM Na2 HPO4, l mM MgSO4, 85 mM NaCl) and transferred to 50-mm NGM plates containing 10 µg/ml 5-fluoro-2’-deoxyuridine with a lawn of E. coli OP50. Worms that failed to move spontaneously or respond to touch were counted as dead. Worms were counted every 2 days [60].
L1 growth assay
The time-course drug treatments were performed using synchronized L1 worms in M9 buffer containing various drugs at 20 o C. Then, worms were transferred to NGM plates and incubated for 4 days. UVC irradiation was performed to synchronized L1 worms, and then worms were incubated for 4 days. After 4 days, the percentage of worms that grew from L1 to adults was calculated, and the half-maximal inhibitory time or dose was obtained by the three-parameter logistic curve using R [55]. At least 200 animals were counted in each condition.
Quantification of germ line apoptosis
Synchronized L1 larvae were cultured on NGM plates until they developed to the young adult stage (2 days). They were harvested with M9 buffer and then treated with various DNA-damaging agents in M9 buffer at 20° C for 24 hours. Worms were then immobilized in M9 buffer containing 20 mM sodium azide and observed using a differential interference contrast (DIC) microscope [33]. At least 150 gonad arms were counted in each condition.
Adult worm drug resistance assay
Synchronized L1 larvae were cultured on NGM plates until they completely developed to the adult stage (4 days). They were harvested with M9 buffer and then treated with various DNA-damaging agents in M9 buffer for 1 hour at 20° C. They were transferred to NGM plates and incubated for 1 day, and the survival rate was then calculated. At least 200 animals were counted in each condition.
AO staining
Synchronized L1 larvae were cultured on NGM plates until they developed completely to the adult stage (4 days). Adult worms were harvested with M9 buffer, and treated with various DNA-damaging agents in M9 buffer for 1 hour at 20° C. Worms were then washed with M9 buffer two times and stained with 5 mg/ml acridine orange (AO) in M9 buffer for 5 minutes. Worms were destained two times with 1 ml of M9 buffer for 10 min, and fixed with phosphate buffered saline (pH 7.4) (PBS) containing 4% paraformaldehyde (PFA). After washing two times with 1ml of M9 buffer, they were observed using fluorescence microscopy with excitation by a 488 nm argon laser [42]. At least 150 animals were counted in each condition.
Feeding RNAi
Knockdown analysis was performed using standard feeding methods [61, 62]. E. coli HT115 (DE3) was transformed with L4440 vectors and cultured overnight in Luria-Bertani (LB) medium containing 25 µg/ml carbenicillin (Cb), 15 µg/ml tetracycline (Tet), and 320 µM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 30 o C. After additional supplementation with Cb, Tet, and IPTG (final 50 µg/ml, 30 µg/ml, and 640 µM, respectively), overnight cultures were seeded on NGM plates (RNAi plates). Worms were primarily cultured on RNAi plates for 3 days. Eggs were then harvested using the alkaline bleach method. The eggs were transferred to fresh RNAi plates and allowed to develop completely to the adult stage (4 days). To confirm the effects of RNAi, reverse transcription-polymerase chain reaction (RT-PCR) was carried out using the primer pairs listed in the Supplementary Table and GoTaq (Promega, USA).
Reporter assay
MAH236 worms cultured on RNAi plates were harvested with M9 buffer and treated with DNA-damaging agents for 7 hours at 20 o C in M9 buffer. Worms were immediately fixed with PBS containing 4% PFA for 10 minutes at 20 o C. After washing two times with M9 buffer, worms were observed using fluorescent microscopy with excitation by a 488 nm argon laser [23]. At least 150 animals were counted in each condition.
Semi-quantitative RT-PCR
In order to determine the expression levels, semi-quantitative RT-PCR was carried out using the primer pairs listed in the Supplementary Table.
Microscopy
DIC microscopy and fluorescence microscopy were performed using a Carl Zeiss LSM510 microscope (Carl Zeiss, Germany).
Statistics
Qualitative data were representative data of at least three experiments. Unless otherwise noted, quantitative data were expressed as the mean ± S.D. The significance of differences was examined using Student’s t-test or the Mann–Whitney U-test. p-values<0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Dr. Elizabeth Nakajima for editing and proofreading. Worms were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), and the National BioResource Project (NBRP). This work was financially supported by Grants-in-Aid for Scientific Research (#24510071, 15K11922) from Ministry of Education, Culture, Sports, Science and Technology of Japan.
CONFLICTS OF INTEREST
The authors declare no competing interests.
- 1. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106(2):302–23. [PubMed]
- 2. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281(41):30305–9. [PubMed] https://doi.org/10.1074/jbc.R600022200.
- 3. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–38. [PubMed]
- 4. DNA mismatch repair system, Classical and fresh roles. FEBS J. 2006;273(8):1609–19. [PubMed]
- 5. Mismatch repair, genetic stability, and cancer. Science. 1994;266(5193):1959–60. [PubMed]
- 6. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol. 2012;30(9):958–64. [PubMed] https://doi.org/10.1200/JCO.2011.39.5590.
- 7. Hepatocellular carcinoma as extracolonic manifestation of Lynch syndrome indicates SEC63 as potential target gene in hepatocarcinogenesis. Scand J Gastroenterol. 2013;48(3):334–51. [PubMed]
- 8. Mismatch repair and DNA damage signalling. DNA Repair (Amst). 2004;3(8-9):1091–101. [PubMed]
- 9. Phosphorylated hMSH6: DNA mismatch versus DNA damage recognition. Mutat Res. 2011;706(1-2):36–45. [PubMed] https://doi.org/10.1016/j.mrfmmm.2010.10.008.
- 10. Exonuclease 1 (Exo1) is required for activating response to S(N)1 DNA methylating agents. DNA Repair (Amst). 2012;11(12):951–64. [PubMed] https://doi.org/10.1016/j.dnarep.2012.09.004.
- 11. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J Biol Chem. 2011;286(12):10411–8. [PubMed] https://doi.org/10.1074/jbc.M110.210989.
- 12. Mismatch repair-mediated G2/M arrest by 6-thioguanine involves the ATR-Chk1 pathway. Biochem Biophys Res Commun. 2004;318(1):297–302. [PubMed]
- 13. The role of DNA mismatch repair in drug resistance. Clin cancer Res. 1998;4(1):1–6. [PubMed]
- 14. Recognition and repair of compound DNA lesions ( base damage and mismatch ) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997;17(2):760–9. [PubMed]
- 15. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999 Sep;4(3):439–44. [PubMed]
- 16. Specific Binding of Human MSH2 MSH6 Mismatch-Repair Protein Heterodimers to DNA Incorporating Thymine- or Uracilcontaining UV Light Photoproducts Opposite Mismatched Bases. J Biol Chem. 1999;274(24):16894–900. [PubMed]
- 17. DNA mismatch repair proteins: potential guardians against genomic instability and tumorigenesis induced by ultraviolet photoproducts. J Invest Dermatol. 2003;121(3):435–40. [PubMed]
- 18. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol Cell. 2012;47(5):669–80. [PubMed]
- 19. Mismatch repair-dependent mutagenesis in nondividing cells. Proc Natl Acad Sci U S A. 2012;109(16):6153–8. [PubMed] https://doi.org/10.1073/pnas.1115361109.
- 20. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70(2):396–417. [PubMed]
- 21. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–56. [PubMed]
- 22. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17(4):443–8. [PubMed] https://doi.org/10.1101/gad.1060703.
- 23. Base excision repair AP endonucleases and mismatch repair act together to induce checkpoint-mediated autophagy. Nat Commun. 2013;4:2674. [PubMed] https://doi.org/10.1038/ncomms3674.
- 24. Role of the hMLH1 DNA Mismatch Repair Protein in Fluoropyrimidine-mediated Cell Death and Cell Cycle Responses Role of the hMLH1 DNA Mismatch Repair Protein in Fluoropyrimidinemediated. Cancer Res. 2001;61(13):5193–201. [PubMed]
- 25. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28(5):1470–9. [PubMed] https://doi.org/10.1128/MCB.01641-07.
- 26. An increase of oxidised nucleotides activates DNA damage checkpoint pathway that regulates post-embryonic development in Caenorhabditis elegans. Mutagenesis. 2014;29(2):107–14. [PubMed] https://doi.org/10.1093/mutage/get067.
- 27. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C elegans. Mol Cell. 2000;5(3):435–43. [PubMed]
- 28. Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc Natl Acad Sci U S A. 2002;99(4):2158–63. [PubMed] https://doi.org/10.1073/pnas.032671599.
- 29. Role of the Msh2 gene in genome maintenance and development in mouse fetuses. Mutat Res Elsevier B.V.; 2012;734(1-2):50–5.. 2002;99(4):2158–63. [PubMed]
- 30. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc Natl Acad Sci U S A. 1997 Apr 1;94(7):3122–7. [PubMed] https://doi.org/10.1073/pnas.94.7.3122.
- 31. The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics]. 1992;131(2):321–32. [PubMed]
- 32. 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nat Rev Cancer. 2003;3(5):330–8. [PubMed]
- 33. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126(5):1011–22. [PubMed]
- 34. DNA polymerase beta is required for efficient DNA strand break repair induced by methyl methanesulfonate but not by hydrogen peroxide. Nucleic Acids Res. 2000;28(16):3040–6. [PubMed] https://doi.org/10.1093/nar/28.16.3040.
- 35. Mitomycins and porfiromycin: chemical mechanism of activation and cross-linking of DNA. Science. 1964;145(3627):55–8. [PubMed]
- 36. Structural and functional properties of CiNTH, an endonuclease III homologue of the ascidian Ciona intestinalis: critical role of N-terminal region. Genes Genet Syst. 2012;87(2):115–24. [PubMed]
- 37. A metalloporphyrin superoxide dismutase mimetic protects against paraquatinduced endothelial cell injury, in vitro. J Pharmacol Exp Ther. 1995;275(3):1227–32. [PubMed]
- 38. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982;151(2):750–5. [PubMed]
- 39. Accommodation and repair of a UV photoproduct in DNA at different rotational settings on the nucleosome surface. J Biol Chem. 2005;280(48):40051–7. [PubMed]
- 40. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell. 2012;22(5):1101–8. [PubMed]
- 41. A spatial and temporal map of C elegans gene expression. Genome Res. 2011;21(2):325–41. [PubMed] https://doi.org/10.1101/gr.114595.110.
- 42. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994 Aug;120(8):2121–9. [PubMed]
- 43. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93(4):519–29. [PubMed]
- 44. Autophagic cell death: the story of a misnomer. Nat Rev Mol cell Biol. 2008;9(12):1004–10. [PubMed] https://doi.org/10.1038/nrm2527.
- 45. The impact of autophagy on cell death modalities. Int J Cell Biol. 2014 Jan;2014:502676. [PubMed] https://doi.org/10.1155/2014/502676.
- 46. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22(12):1931–7. [PubMed]
- 47. Loss of ATM sensitizes against O6-methylguanine triggered apoptosis, SCEs and chromosomal aberrations. DNA Repair (Amst). 2004;3(4):359–68. [PubMed]
- 48. Alkylation damage causes MMR-dependent chromosomal instability in vertebrate embryos. Nucleic Acids Res. 2008;36(12):4047–56. [PubMed] https://doi.org/10.1093/nar/gkn341.
- 49. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16(8):819–24. [PubMed]
- 50. The atm-1 gene is required for genome stability in Caenorhabditis elegans. Mol Genet Genomics. 2012;287(4):325–35. [PubMed] https://doi.org/10.1007/s00438-012-0681-0.
- 51. DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Dev Cell.2011;20(3):353–63.. 2012;287(4):325–35. [PubMed]
- 52. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22(4):501–10. [PubMed] https://doi.org/10.1016/j.molcel.2006.04.023.
- 53. A conserved pathway to activate BRCA1dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25(10):2178–88. [PubMed] https://doi.org/10.1038/sj.emboj.7601102.
- 54. Reduced expression of hMLH1 and hMSH2 gene products in highgrade hepatocellular carcinoma. Acta Med Okayama. 2001;55(2):65–71. [PubMed]
- 55. Purification and characterization of Caenorhabditis elegans NTH, a homolog of human endonuclease III: Essential role of N-terminal region. DNA Repair (Amst). 2009;8(7):844–51. [PubMed]
- 56. A novel protein, MAPO1, that functions in apoptosis triggered by O6-methylguanine mispair in DNA. Oncogene. 2009;28(8):1142–50. [PubMed]
- 57. Caffeine acutely activates 5’adenosine monophosphate-activated protein kinase and increases insulin-independent glucose transport in rat skeletal muscles. Metabolism.2009;58(11):1609–17.. 2009;28(8):1142–50.
- 58. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–9. [PubMed]
- 59. Extension of lifespan and protection against oxidative stress by an antioxidant herb mixture complex (KPG7) in Caenorhabditis elegans. J Clin Biochem Nutr. 2013;53(2):81–8. [PubMed] https://doi.org/10.3164/jcbn.13-11.
- 60. Oxidation resistance 1 (OXR1) is essential for protection against oxidative stress and participates in the regulation of aging in Caenorhabditis elegans. Free Radic Res. 2014;48(8):919–28. [PubMed]
- 61. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–21. [PubMed]
- 62. Specific interference by ingested dsRNA. Nature. 1998;395(6705):854. [PubMed]
Last Modified: 2016-06-03 03:54:52 EDT
PII: 70
