Genes & Cancer
Hexokinase II inhibitor, 3-BrPA induced autophagy by stimulating ROS formation in human breast cancer cells
Qianwen Zhang1, Yuanyuan Zhang1, Pei Zhang1, Zhenhua Chao1, Fei Xia1, Chenchen Jiang2, Xudong Zhang2, Zhiwen Jiang1, Hao Liu1
1Faculty of pharmacy, Bengbu Medical College, Bengbu, Anhui, P. R. China
2School of Medicine and Public Health, Faculty of Health, University of Newcastle, NSW, Australia
Correspondence to: Hao Liu, email: [email protected]
Keywords: 3-BrPA, CQ, Autophagy, Apoptosis, Necroptosis, ROS
Received: April 22, 2014
Accepted: May 15, 2014
Published: May 16, 2014
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT:
Hexokinase II (HKII), a key enzyme of glycolysis, is widely over-expressed in cancer cells. 3-bromopyruvate (3-BrPA), an inhibitor of HK II, has been proposed as a specific antitumor agent. Autophagy is a process that regulates the balance between protein synthesis and protein degradation. Autophagy in mammalian systems occurs under basal conditions and can be stimulated by stresses, including starvation, oxidative stress. Therefore, we hypothesized that 3-BrPA could induce autophagy. In the present study, we explored the mechanism of 3-BrPA and its combined action with chloroquine. Our results demonstrate that in MDA-MB-435 and in MDA-MB-231 cells, 3-BrPA induces autophagy, which can be inhibited by chloroquine. Furthermore, the combined treatment synergistically decreased the number of viable cells. Interestingly, the combined treatment triggered apoptosis in MDA-MB-435 cells, while it induced necroptosis in MDA-MB-231 cells. ROS mediated cell death when 3-BrPA and CQ were co-administered. Finally, CQ enhanced the anticancer efficacy of 3-BrPA in vivo. Collectively, our results show that 3-BrPA triggers autophagy, increasing breast cancer cell resistance to 3-BrPA treatment and that CQ enhanced 3-BrPA-induced cell death in breast cancer cells by stimulating ROS formation. Thus, inhibition of autophagy may be an innovative strategy for adjuvant chemotherapy of breast cancer.
INTRODUCTION
Cancer cells undergo increased aerobic glycolysis, increasingly relying on this metabolic pathway to generate ATP to sustain elevated proliferation. That an impaired glucose metabolism leads to lactic acid secretion in the presence of oxygen was first described by Warburg in the 1920s [1]. This increased aerobic glycolysis has since been observed in many tumor cells. Consequently, specific drugs designed to interfere with energy-producing pathways of many cancer types have been investigated [2, 3], such as the glycolytic inhibitors 2-deoxyglucose [4], and 3-BrPA [5]. 3-BrPA is an analogue of pyruvate with high tumor selectivity [6, 7]. In mammals, there are four hexokinase isoforms, HK1, HK2, HK3, and HK4, encoded by separate genes. HK2 is expressed at relatively high level only in skeletal muscles, adipose tissues, and heart. Despite its absence or low expression in the majority of adult normal cells, HK2 is widely over-expressed in many cancer cells [8]. Patra’s group showed that HK2 expression is dramatically elevated in tumors derived from mouse models of lung and breast cancer [9]. Cecilia’s group found that HK I and II inhibition by metformin could modify glucose metabolism in triple-negative breast cancer both in cultured cells and xenograft models [10]. So it seemed feasible that HK2 could be a selective therapeutic target for cancer. 3-BrPA can not only inhibit HKII and prevent glucose from entering the glycolytic pathway [11], but the compound can also induce cell death by activating the mitochondrial pathway of apoptosis or necrosis [12, 13].
Programmed cell death is executed through specific intracellular biochemical pathways. Apoptosis, autophagy, necrosis and/or necroptosis are three common forms of programmed cell death distinguished according to their morphological, enzymological and functional criteria. Apoptosis is executed by a group of intracellular cysteine proteases, namely caspases. Necrosis, on the other hand, is associated with organelle swelling, cytoplasmic membrane breakdown, and ensuing inflammation responses [14]. Necroptosis is a regulated necrotic cell death triggered by broad caspase inhibition and is characterized by necrotic cell death morphology and the activation of autophagy [15]. Receptor-interacting protein kinase 1 (RIP1) kinase activity is then crucial for this pathway, which may also be mediated via the Fas, TNF or TNF-related apoptosis-inducing ligand (TRAIL) death receptors [16]. Autophagy is a major intracellular pathway for the degradation and recycling of unused but long-lived proteins, damaged organelles, and even invasive pathogens [17, 18]. Some studies have highlighted the importance of autophagy in several organs, including the brain, heart, hematopoietic cells, and the kidney [19, 20, 21, 22]. For instance, systemic autophagy-knockout mice die within one day after birth [23]. Although autophagy is important for normal cell function and survival, it is also used by tumor cells and may be therapeutically counterproductive [24, 25]. Activation of autophagy has been suggested to promote cell survival [26], so a combination chemotherapeutic treatment with an autophagy inhibitor could promote tumor cell death [27]. Chloroquine (CQ) is an anti-malarial drug, as well as a well-known lysosomotropic agent that inhibits late-stage autophagy [28]. An analogue of CQ, hydroxychloroquine (HCQ), has also been shown to have antitumor properties [29]. HCQ promotes tumor cell death though either p53 activation or alkylation in a mouse model of c-Myc-driven lymphoma [30]. However, the precise mechanism by which autophagy inhibition promotes cancer cell death remains to be determined, particularly with respect to glycolytic inhibitors and the definition of cancer susceptibility to autophagy inhibition.
The role of autophagy in 3-BrPA-induced cell death in human breast cancer cells has not, to our knowledge, been explored. Thus, we investigated whether autophagic machinery could be activated in breast cancer cells after 3-BrPA treatment and whether inhibition of autophagy enhanced 3-BrPA-mediated cell death. Furthermore, we demonstrated that ROS formation increased cell sensitivity to death via autophagy inhibition. Finally, we demonstrated that inhibition of autophagy enhanced 3-BrPA-mediated cell death in vivo.
RESULTS
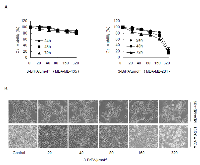
The effects of 3-BrPA on cell proliferation in breast cancer cell lines MDA-MB-435 and MDA-MB-231
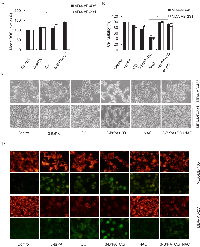
As an analogue of pyruvate, 3-BrPA can induce cell death in certain tumor cell lines. To investigate 3-BrPA’s potential to inhibit cell growth in breast cancer cells (MDA-MB-231, MDA-MB-435), cell viability was measured with an MTT assay after incubation of each cell line with 3-BrPA for various periods. As shown in Figure 1A, 3-BrPA (0–320 μM) reduced MDA-MB-231 cell growth in a dose- and time- dependent manner. 3-BrPA had little effect on MDA-MB-435 cell growth (Figure 1A). As visualized with an inverted microscope, viable MDA-MB-231 cells decreased significantly and dose-dependent manner after 24 h of 3-BrPA treatment. Most cells shrank and became rounded before detaching from the culture plates (Figure 1B), whereas viable MDA-MB-435 cells only modestly decreased. These data prompted us to explore the unique effects of 3-BrPA on MDA-MB-435 and MDA-MB-231 cells.
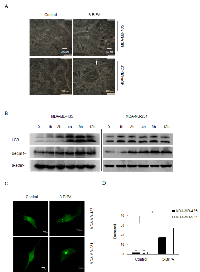
3-BrPA induces autophagy in breast cancer cells
First electron microscopy (EM) was used to visualize cell morphology after both cell lines were treated with 3-BrPA. As shown in Figure 2A, 3-BrPA treatment increased the presence of autophagosomes filled with debris in both cell lines; only a few vacuoles were observed in control cells. Autophagy-specific markers such as microtubule-associated protein 1 LC3, Beclin-1 were used to quantify autophagy with immunoblot analysis. As shown in Figure 2B, conversion of LC3 I/II and up-regulation of Beclin-1 suggested increased formation of autophagosomes in a time-dependent manner in breast cancer cells. Induction of autophagy by 3-BrPA was then examined by imaging the cellular distribution of GFP-LC3, a fusion construct of green fluorescent protein with LC3. In control cells, GFP-LC3 puncta were mainly distributed to the cytosol, indicating a low level of autophagy under these conditions (Figure 2C, left panel). In contrast, cells incubated with 3-BrPA for 12 h revealed many autophagosomes with accumulated GFP-LC3 (presumably as GFP-LC3-II). Quantification data for puncta per cell are depicted in Figure 2D. These data suggest that 3-BrPA induces a complete autophagic response in breast cancer cells.
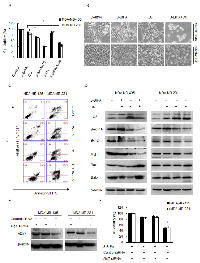
Autophagy inhibition enhances 3-BrPA-induced cell death
To investigate the role of autophagy in cell death, we inhibited autophagy with CQ or 3-Methyladenine (3-MA) and measured cell death. As expected, a significant increase in 3-BrPA-induced cell death was observed in breast cancer cells after autophagy was inhibited with CQ or 3-MA (Figure 3A). As viewed with an inverted microscope, viable MDA-MB-231 and MDA-MB-435 cells were significantly decreased after 3-BrPA plus CQ treatment for 12 h. The majority of cells shrank and became round before detaching from the culture plates (Figure 3B). Moreover, annexin V-propidium iodide staining revealed a significant increase in cell death (defined as annexin V+ and PI V+cells) after 3-BrPA plus CQ treatment (Figure 3C).
To ascertain whether 3-BrPA plus CQ affects autophagy, we measured expression of autophagy-related proteins using western blot. Accumulation of LC3-II indicates high flux through the autophagy pathway, 3-BrPA increased turnover and clearance of LC3-positive autophagosomes and LC3-II as indicated by western blot and increased autophagic flux blocked was with CQ. Accordingly, the concentration of Beclin1 was reduced by 3-BrPA in the presence of CQ (Figure 3D). Examining apoptosis-related proteins, we discovered that 3-BrPA plus CQ treatment significantly inhibited anti-apoptotic Bcl-2 and MCl-1 expression and enhanced pro-apoptotic Bax and Bak expression (Figure 3D). Knockdown of Atg7 siRNA also sensitized breast cancer cells to 3-BrPA, indicating that autophagy was responsible for 3-BrPA resistance (Figure 3E and 3F). These data suggest that autophagy is a mechanism of resistance to 3-BrPA that could be reversed by genetic blockade of autophagy initiation or pharmacologic blockade of cargo degradation.
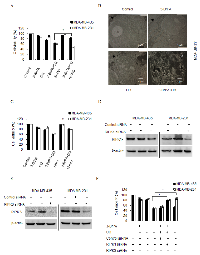
3-BrPA plus CQ treatment induces RIPK1-dependent apoptosis in MDA-MB-435 cells and RIPK1/3-dependent necroptosis in MDA-MB-231 cells
To confirm the cell death type induced by the combination of 3-BrPA and CQ, we used a broad spectrum caspase inhibitor, z-VAD-fmk to rescue cells from death induced by 3-BrPA plus CQ treatment. As shown in Figures 4A, an MTT assay revealed that z-VAD-fmk rescued MDA-MB-435 cells, but exacerbated cell death in MDA-MB-231 cells. Using EM, we observed that 3-BrPA plus CQ-treated MDA-MB-231 cells had ruptured plasma membranes, indicating necrosis (Figure 4B). Death may also occur by a programmed form of necrosis, necroptosis, which requires RIPK1 and RIPK3 activation by death receptors [31] or other death receptor-independent mechanisms [32]. To assess whether 3-BrPA plus CQ treatment induced necroptosis, MDA-MB-231 cells were treated with 3-BrPA plus CQ with/without the specific RIPK1 and necroptosis inhibitor necrostatin-1 (Nec-1). Nec-1 alone had no effect on cells, but dramatically restored cell survival by after treatment with 3-BrPA plus CQ in MDA-MB-231 cells, and it also had the same effect on MDA-MB-435 cells (Figures 4C). Knockdown of RIPK1 in both cell lines (Figure 4D) also suppressed cell death and enhanced cell viability (Figure 4F). Similarly, MDA-MB-231 cells treated with a validated siRNA against RIPK3 (Figure 4E) were significantly more viable after treatment with 3-BrPA plus CQ, while knockdown of RIPK3 had no effect on MDA-MB-435 cells (Figure 4F). These data suggest that 3-BrPA and CQ induce two different cell death types in MDA-MB-435 cells and MDA-MB-231 cells, and both death pathways require RIPK1.
ROS production causes cell death with 3-BrPA and autophagy inhibition
Next, to elucidate the mechanism which leads to the initiation of 3-BrPA-related autophagy, we quantified intracellular ROS in breast cancer cells after a 12 h incubation with 3-BrPA, CQ, or a combination of both drugs. As shown in Figure 5A, ROS generation was markedly increased in cells treated with 3-BrPA plus CQ compared to cells treated with 3-BrPA alone. Because elevated ROS occurred in cells with suppressed autophagy, we investigated whether ROS inhibition influenced 3-BrPA-plus-CQ-mediated cell death. ROS formation was inhibited with the ROS scavenger N-acetyl cysteine (NAC) and cell viability was measured with an MTT assay. Cell morphology was observed and photographed under an inverted microscope (Figures 5B, 5C). Our data indicate that cell sensitization to 3-BrPA- and CQ-induced completely blocked cell death when ROS formation was inhibited. In addition, the mitochondrial membrane potential (MMP), an indicator of ROS generation, was detected in these cells using JC-1 [33]. Higher MMP was observed in the control culture group compared to the dually treated groups (Figure 5D). With co-treatment with 3-BrPA and CQ, JC-1 revealed higher green fluorescence, while adding NAC makes MMP turn to be higher again. (Figure 5D).
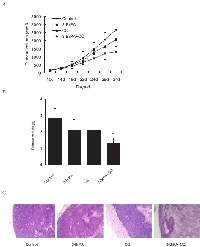
CQ enhanced anti-tumor efficacy of 3-BrPA in nude mice
To ascertain whether a combination of 3-BrPA and CQ could suppress tumor growth, MDA-MB-231 xenografts were examined. Tumors continued to grow in xenografted mice treated with vehicle, CQ, or 3-BrPA, or a combination of CQ and 3-BrPA. The combination of CQ plus 3-BrPA prevented tumor growth (Figure 6A). After treatment ended, mice were sacrificed, and tumors were excised and evaluated. Tumor weights of mice treated with vehicle, CQ, and 3-BrPA alone were greater than those from mice treated with CQ plus 3-BrPA (Figure 6B). Hematoxylin and eosin (HE) staining indicated greater necrotic areas in CQ plus 3-BrPA-treated mice. Thus, CQ enhanced the anti-tumor efficacy of 3-BrPA in vivo.
DISCUSSION
The present study demonstrates that exposure of breast cancer cells to 3-BrPA induces autophagy and that CQ enhances the anti-tumor efficacy of 3-BrPA, both in vitro and in vivo. The lethal effect of 3-BrPA in combination with CQ is initially triggered by the development of ROS-dependent autophagy induced by 3-BrPA, and then inhibition of autophagy and subsequent induction of mitochondrial depolarization and more ROS production, resulting in two different forms of cell death in MDA-MB-435 cells and MDA-MB-231 cells.
The roles of autophagy within the context of cancer are controversial. One view is that autophagy suppresses tumor progress; another view is that autophagy is a pro-survival pathway. Inactivation of the autophagy-specific gene, Beclin-1, produces significantly higher frequencies of spontaneous malignancies in mice [34]. Muhammad’s group showed that palmitic acid triggers Ca2+-dependent autophagy, which results in programmed necrotic death (necroptosis) in endothelial cells [35]. However, autophagy may also be a pro-survival pathway assisting tumor cells to endure metabolic stress under nutrient/oxygen-deficient conditions and resist death triggered by chemotherapeutic agents. Xu’s group showed that accumulation of autophagosomes in breast cancer cells induced TRAIL resistance through downregulation of surface expression of death receptors 4 and 5 [36]. Wang’s laboratory reported that reduced miRNA MIR23B increases ATG12 and autophagy to promote radioresistance in pancreatic cancer cells [37]. An analysis of the transcriptional status of ATG12 in > 50 breast cancer cell lines suggested that the ATG12 transcript is commonly upregulated in trastuzumab-unresponsive HER2-overexpressing breast cancer cells [38].
Here, we report that 3-BrPA had different effects in two breast cancer cell lines (Figure 1) although autophagy occurred after drug treatment in both lines (Figure 2). Autophagy inhibitors CQ and 3-MA used during 3-BrPA treatment revealed that CQ and 3-MA enhanced 3-BrPA-induced cell death in both cell lines (Figure 3A, 3B and 3C). Then, siRNA-mediated knockdown of Atg7 sensitized cells to 3-BrPA treatment (Figure 3E, 3F). Thus, autophagy played a protective role during 3-BrPA treatment.
Autophagy can prevent apoptosis [39], necrosis [40], and necroptosis [41]. We used z-VAD-fmk to rule out the contribution of pan-caspases in 3-BrPA plus CQ-induced cell death. z-VAD rescued 3-BrPA plus CQ-induced cell death in MDA-MB-435 cells but exacerbated cell death in MDA-MB-231 cells. Furthermore, MDA-MB-231 underwent necrotic cell death after 3-BrPA plus CQ treatment, as evidenced by EM. Thus, necrosis is the predominant mode of cell death in this line. Finally, Nec-1 rescued 3-BrPA plus CQ-induced cell death in MDA-MB-231 and MDA-MB-435 cells. These findings strongly suggest that 3-BrPA plus CQ induce necroptosis in MDA-MB-231 cells. RIPK1 is a multi-functional protein also known to mediate NF-κB and caspase-8 activation in response to TNF-α[42]. The pro-apoptotic function of RIPK1 is the formation of a caspase-8 activation complex in response to TNF receptor I activation [43]. RIPK1 was also reported to be important for triggering necroptosis [44]. In our study, knockdown of RIPK1 rescued both two cells from 3-BrPA plus CQ-induced cell death. In addition, knockdown of other known contributors to programmed necrosis, such as RIPK3, only rescued MDA-MB-231 cells from 3-BrPA-induced cell death. Interestingly, 3-BrPA plus CQ induces apoptosis in MDA-MB-435 cells, but induces necroptosis in MDA-MB-231 cells, and all require RIPK1. These data suggest a potential for novel strategies for the treatment of breast cancer.
Autophagy induction after mitochondrial damage can lead to ROS production [45]. ROS, at the physiological level, are important for a variety of cellular programs of physiological and pathological conditions. However, aberrantly high ROS are intimately associated with disease and are commonly observed in cancer [46]. ROS regulate autophagy by multiple mechanisms that involve direct effects on Atg protein Atg4 [47], affecting the activity of the master autophagy regulator mTORC1 [48]. ROS are also implicated in autophagy induction in cancer therapy [49], suggesting that ROS are important with respect to the mechanism of actions of cancer chemotherapeutics. Deregulation of ROS formation is associated with cancer initiation, progression, and drug resistance. In our study, inhibition of autophagy led to an increase in ROS formation and blocking ROS accumulation with NAC rescued 3-BrPA plus CQ-induced cell death (Figure 5A, 5B, 5C), suggesting that ROS accumulation is an important mechanism in the sensitization of cells to death when autophagy is suppressed.
In summary, 3-BrPA-induced autophagy in breast cancer cells may function as a resistance mechanism against cell death. Then, inhibition of autophagy could be a novel strategy for breast cancer adjuvant therapy. Although detailed mechanisms driving ROS generation and autophagy in our model are unclear, our data will inform future studies in cancer cells treated with 3-BrPA.
MATERIALS AND METHODS
Ethics statement
Tumor assays in mice were performed using Institutional Animal Care and Use and proved by the Committee on the Ethics of Animal Experiments of the Bengbu Medical College (Permit Number: 013). Mice were monitored for overall health in response to drug treatment. All efforts were made to minimize suffering during the experiment.
Materials
Cell culture materials were obtained from Invitrogen and fetal calf serum was from Gibco. 3-bromopyruvate, chloroquine diphosphate, 3-methyladenine (3-MA), 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT), Nec1 were purchased from Sigma-Aldrich. z-VAD-fmk was purchased from Calbiochem. JC-1 and Annexin V-FICT/PI assay were purchased from KeyGEN BioTECH (China). GFP-LC3 plasmid was purchased from GeneCopoeia. The following antibodies were used: LC3, Bclin-1 (MBL), Bax, Bak, Bcl-2, Mcl-1 (ProteinTech), Atg7 (Beyotime), RIPK1 (Santa cruz), RIPK3 (Cell Signaling).
Cell Culture
MDA-MB-435 and MDA-MB-231 cells were purchased from Cell Bank (Chinese Academy of Sciences). Cells were grown in DMEM containing 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, and were maintained at 37 °C in a humidified atmosphere of 5% CO2 in air.
SiRNA Design and Validation
siRNA against ATG7 (GGUCAAAGGACGAAGAUAATT UUAUCUUCGUCCUUUGACCTT), RIPK1 (GCAAAGACCUUACGAGAAUTT AUUCUCGUAAGGUCUUUGCTT) and RIPK3 (CCAGUGACGUCUACAGCUUTT AAGCUGUAGACGUCACUGGTT) were designed by Gene Pharma. Knockdown efficiency of individual siRNA was validated by western blot analysis. All siRNAs were obtained from Gene Pharma.
RNA interference
Cells were grown on 30-mm glass coverslips to 75% confluence and transfected with either siRNA or plasmid using the LipofectamineTM 2000 transfection reagent from Invitrogen. 70 pmol of the respective siRNA(s) were mixed with transfection reagent in 0.5 ml of opti-MEM (Invitrogen) without FCS and incubated at room temperature for 20 min. Cells were incubated for 24 h and the medium was exchanged with complete culture medium. 48 h post-transfection, cells were incubated with drugs and viability was assessed by MTT assay.
Plasmid transfection and confocal microscopy analysis
For overexpression of GFP-LC3 cells were transfected with 1 ml of serum-free opti-MEM containing 2 μg of plasmid DNA and 5 μl of LipofectamineTM 2000. The medium was complemented after 1 h with 1ml of full culture medium. Cells were incubated for 6 h and the medium was replaced by complete culture medium. 48 h post-transfection, cells were incubated with drugs. The treated cells were fixed with 4% paraformaldehyde and examined by a confocal microscopy (Nikon D-Eclipse-C1).
Electron microscopic detection of autophagosome
Cells were collected and fixed with 3% glutaraldehyde and 2% paraformaldehyde in 0.1 M PBS buffer (pH 7.4) for 30 min, postfixed with 1% osmium tetroxide for 1.5 h, washed and stained in 3% aqueous uranyl acetate for 1 h then dehydrated in an ascending series of ethanol and acetone, and embedded in Araldite. Ultrathin sections were cut on a Reichert ultramicrotome, double stained with 0.3% lead citrate and examined on OLYMPUS JEOL electron microscope.
MTT Assay
Cells were plated in a 96-well plate and allowed to adhere overnight before drug treatment. After the indicated treatments, cells were incubated with MTT at a final concentration of 0.5 mg/L. After 3-4 h, the medium was removed, and the cells were dissolved in DMSO 150 μL each well and the blue dye was allowed to dissolve for 30 min at room temperature. The absorbance was subsequently measured at 570 nm using a microplate reader (BioTek). Data were normalized to respective controls and represented as percent viability of the controls.
AnnexinV-FITC/PI Staining
The detection was performed by the AnnexinV-FITC Apoptosis Detection Kit (KeyGEN BioTECH). Briefly, cells were seeded in 6-well plates and incubated for 12 h and then exposed to drugs. After treatment, approximately 1×105 cells were harvested and washed with warm PBS prior to stain with Annexin V-FITC and PI according to the manufacturer’s instructions. After 20 min of incubation, cells were analyzed by flow cytometry (BD, Accuri C6).
Western Blotting
Cells were washed twice with ice-cold PBS and lysed. The protein concentration was measured using a BCA protein assay kit (Beyotime). An equal amount of protein were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% nonfat dry milk in PBS containing 0.05% Tween 20 (Sigma) for 4 h and then incubated with the primary antibody at 4 °C overnight. The following day, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody for 2 h. The membrane was further developed with the ECL plus western blotting detection system (BioTek).To control the equal amount of protein loading, membrane was reprobed with β-actin antibody.
Mitochondrial Membrane Potential (MMP) Analysis
The detection was performed by fluorescence microscope using JC-1 staining according to the manufacturer’s instructions. Briefly, following drug treatment, cells were incubated with 10 μM JC-1 for 30 min at 37 °C in the dark. Then cells were detected by fluorescence microscope (Olympus, 1X71).
Measurement of reactive oxygen species (ROS)
Intracellular ROS levels were measured using dihydroethidium (DHE), a cell-permeable indicator for ROS generation. Following drug incubation, 5 μM DHE added to the media and incubated 30 minutes in the dark. Then cells were harvested, washed with PBS and detected by flow cytometry. The results were analyzed by Cell Quest analysis software.
In vivo tumor experiment
The nude mice (5-6 weeks) used in these studies were obtained from Beijing vitalriver and weighed 20–25 g at the time of tumor implantation. The mice were kept under a 12:12 h light–dark cycle, at 24 ± 2 °C and fed with clean food and water. Human MDA-MB-231 cells (107 cells/ml) were inoculated subcutaneously to form tumors. Mice with tumors (100-200 mm3) assorted to four groups (5 mice/group). Vehicle (0.9% NS) or CQ (40mg/kg/d, 24 days) or 3-BrPA (5mg/kg/d, 24 days) alone or in combination was administrated intraperitoneally. Tumor growth was monitored every three days by two-dimensional measurements of individual tumors for each mouse. Tumor volume was calculated using the formula: length×width2/2. After treatment ended, mice from each group were sacrificed. Tumors were excised, calculated and fixed in 4 % formalin solution, embedded in paraffin, and then stained with hematoxillin-eosin (H&E).
Statistical analysis
Statistical analyses were performed with Concise Statistics (CS 14.0) software. The data presented were mean ±standard deviation (SD). Data were compared using Student’s t test. p< 0.05 was considered significant.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81000992, 81072207) and the Natural Science Foundation of Anhui Province (090413135), Education Department of Anhui Natural Science Research Key Project China (KJ2012A202).
Authors’ contributions
QWZ conducted the experiments and wrote the manuscript. YYZ, PZ, FX, ZHC participated in the design and coordination of study. CCJ, ZWJ, HL conceived of the study, participated in the design and helped to draft the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that they have no competing interests.
- 1. On respiratory impairment in cancer cells. Science. 1956, 124:267-9. [PubMed]
- 2. Energy metabolism in tumor cells. FEBS. 2007; 274:1393-418. [PubMed]
- 3. The cancer cell’s ‘power plants’ as promising therapeutic targets an overview. Bioenerg Biomembr. 2007; 39:1-12.
- 4. Cytotoxicity, radiosensitization, and chemosensitization of tumor cells by 2-deoxy-D-glucose in vitro. Cancer Res Ther. 2009; 5:S27-S31. [PubMed]
- 5. H. and Pedersen PL. Hexokinase-2 bound to mitochondria: cancer’s stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin Cancer Biol. 2009; 19:17-24.
- 6. 3-bromopyruvate kills cancer cells in animals. Lancet Oncol. 2002; 3:524.
- 7. Novel therapy for liver cancer: direct intra arterial injection of a potent inhibitor of ATP production. Cancer Res. 2002; 62:3909-13. [PubMed]
- 8. Hexokinase 2 as oncotarget. Oncotarget. 2013; 4:1862-3. [PubMed] https://doi.org/10.18632/oncotarget.1563.
- 9. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013; 24:213-28. [PubMed] https://doi.org/10.1016/j.ccr.2013.06.014.
- 10. Direct inhibition of hexokinase activity by metformin at least partially impairs glucose metabolism and tumor growth in experimental breast cancer. Cell Cycle. 2013; 12:3490-9. [PubMed] https://doi.org/10.4161/cc.26461.
- 11. Glucose catabolism in the rabbit VX2 tumor model for liver cancer: characterization and targeting hexokinase. Cancer Lett. 2001; 173:83-91. [PubMed]
- 12. Inhibition of glycolysis in cancer cells: a novel strategy to overcome compound resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Re. 2005; 65:613-21.
- 13. Apoptosis-inducing antitumor efficacy of hexokinase II inhibitor in hepatocellular carcinoma. Mol Cancer Ther. 2007; 6:2554-62. [PubMed]
- 14. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochimica. Et. Biophysica. Acta. 2006; 1757: 1371–87. [PubMed]
- 15. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005; 1: 112–9. [PubMed]
- 16. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000; 1: 489 –95. [PubMed]
- 17. Autophagy as a regulated pathway of cellular degradation. Science. 2000; 90:1717-21. [PubMed]
- 18. Autophagy: renovation of cells and tissues. Cell. 2011; 147:728-41. [PubMed]
- 19. Loss of autophagy in the central nervous system causes neurodeg eneration in mice. Nature. 2006; 441:880-4.
- 20. The role of autophagy in cardiomy ocytes in the basal state and in response to hemody namic stress. Nat Med. 2007; 13:61924. [PubMed]
- 21. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011; 208:455-67. [PubMed] https://doi.org/10.1084/jem.20101145.
- 22. Autophagy guards against cisplatin-induced acute kidney injury. Am J Pathol. 2012; 180:517-25. [PubMed]
- 23. The role of autophagy during the early neonatal starvation period. Nature. 2004; 432:1032-6. [PubMed]
- 24. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008; 4:563-66. [PubMed]
- 25. Autophagy in tumorigenesis and energy metabolism:friend by day, foe by night. Curr Opin Genet Dev. 2011; 21:113-19. [PubMed] https://doi.org/10.1016/j.gde.2010.12.008.
- 26. Parkin is recruited sel -ectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008; 183:795-803. [PubMed] https://doi.org/10.1083/jcb.200809125.
- 27. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012; 12:401-10. [PubMed] https://doi.org/10.1038/nrc3262.
- 28. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009; 625:220-33. [PubMed]
- 29. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006; 144:337-43. [PubMed]
- 30. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007; 117:326-36. [PubMed] https://doi.org/10.1172/JCI28833.
- 31. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008; 135:1161-3. [PubMed]
- 32. Nitration of the mitochondrial complex Ⅰsubunit NDUFB8 elicits RIP1-and RIP3-mediated necrosis. Free Radic Biol Med. 2010; 48:306-17. [PubMed] https://doi.org/10.1016/j.freeradbiomed.2009.11.001.
- 33. From death receptor to reactive oxygen species and c-Jun N-terminal protein kinase: the receptorinteracting protein 1 odyssey. Immunol Rev. 2007; 220:821. [PubMed]
- 34. Promotion of Tumorigenesis by Heterozygous Disruption of the Beclin 1 Autophagy Gene. J Clin Invest. 2003; 112:1809-20. [PubMed] https://doi.org/10.1172/JCI20039.
- 35. Graier, and Roland Malli. Inhibition of Autophagy Rescues Palmitic Acid-induced Necroptosis of Endothelial Cells. J Biol Chem. 2012; 287:21110-20.
- 36. Accumulation of autophagosomes in breast cancer cells induces TRAIL resistance through downregulation of surface expression of death receptors 4 and 5. Oncotarget. 2013; 4:1349-64. [PubMed] https://doi.org/10.18632/oncotarget.1174.
- 37. MicroRNA 23b Regulates Autophagy Associated with Radioresistance of Pancreatic Cancer Cells. Gastroenterology. 2013; 145:113343.e12. [PubMed]
- 38. Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment. Oncotarget. 2012; 3:1600-14. [PubMed] https://doi.org/10.18632/oncotarget.742.
- 39. Antiestrogen-binding site ligands induce autophagy in myeloma cells that proceeds through alteration of cholesterol metabolism. Oncotarget. 2013; 4:911-22. [PubMed] https://doi.org/10.18632/oncotarget.1066.
- 40. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005; 25:1025-40. [PubMed] https://doi.org/10.1128/MCB.25.3.1025-1040.2005.
- 41. Autophagy Suppresses RIP Kinase-Dependent Necrosis Enabling Survival to mTOR Inhibition. PLoS One. 2012; 7:e41831. [PubMed] https://doi.org/10.1371/journal.pone.0041831.
- 42. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996; 4:387–96. [PubMed]
- 43. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008; 133,693–703. [PubMed]
- 44. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010; 11,700-14. [PubMed]
- 45. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007; 120:4155-66. [PubMed]
- 46. ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007; 8:813-24. [PubMed]
- 47. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007; 26:1749-60. [PubMed] https://doi.org/10.1038/sj.emboj.7601623.
- 48. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011; 12:21-35. [PubMed] https://doi.org/10.1038/nrm3025.
- 49. STAT3 transcriptional factor activated by reactive oxygen species induces IL6 in starvation-induced autophagy of cancer cells. Autophagy. 2010; 6:1125-38. [PubMed]
Last Modified: 2016-06-08 14:52:53 EDT
PII: 9
